pDsRed-Express-C1
pDsRed-Express-C1
编号 | 载体名称 |
北京华越洋生物VECT6015 | pDsRed-Express-C1 |
pDsRed-Express-C1载体基本信息
载体名称: | pDsRed-Express-C1 |
质粒类型: | 哺乳动物细胞表达载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | CMV |
载体大小: | 4680 bp |
5' 测序引物及序列: | 5'd[AGCTGGACATCACCTCCCACAACG] |
3' 测序引物及序列: | -- |
载体标签: | 红色荧光蛋白DsRed-Express(N-端) |
载体抗性: | 卡那霉素 |
筛选标记: | 新霉素(Neomycin) |
克隆菌株: | DH5α, HB101 |
宿主细胞(系): | 常规细胞系,293、CV-1、CHO等 |
备注: | pDsRed-Express-C1载体是红色荧光报告载体,表达N端DsRed-Express融合蛋白; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
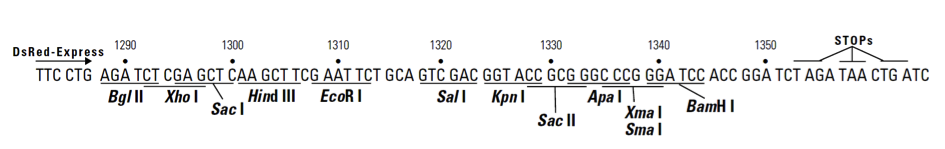
pDsRed-Express-C1载体质粒图谱和多克隆位点信息
pDsRed-Express-C1载体描述
pDsRed-Express-C1 is a mammalian expression vector that encodes DsRed-Express, a variant of Discosoma sp. red fluorescent protein (DsRed; 1). DsRed-Express contains nine amino acid substitutions, which improve the solubility of the protein and reduce the time from transfection to detection of red fluorescence (2). In addition, these substitutions reduce the level of residual green emission (2). When DsRed-Express is expressed in mammalian cell cultures, redemitting cells can be detected by either fluorescence microscopy or flow cytometry 8–12 hours after transfection (DsRed-Express excitation and emission maxima = 557 nm and 579 nm, respectively). Although DsRed-Express most likely forms the same tetrameric structure as wild-type DsRed, DsRed- Express displays a reduced tendency to aggregate (2). The DsRed-Express coding sequence is human codon-optimized for high expression in mammalian cells (3). The multiple cloning site (MCS) in pDsRed-Express-C1 is positioned between the DsRed-Express coding sequence and the SV40 polyadenylation signal (SV40 poly A). Genes cloned into the MCS will be expressed as fusions to the C-terminus of DsRed-Express if they are in the same reading frame as DsRed-Express and there are no intervening stop codons. A Kozak consensus translation initiation site upstream of DsRed-Express increases the translation efficiency in eukaryotic cells (4). SV40 poly A signals downstream of the MCS direct proper processing of the 3' end of mRNA transcripts. The vector backbone also contains an SV40 origin for replication in mammalian cells expressing the SV40 T-antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin resistance cassette (Neor)—consisting of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene—allows stably transfected eukaryotic cells to be selected using G418. A bacterial promoter upstream of this cassette expresses kanamycin resistance in E. coli.
pDsRed-Express-C1 can be used to construct fusions to the C-terminus of DsRed-Express. If a fusion construct retains the fluorescent properties of the native DsRed-Express protein, its expression can be monitored by flow cytometry and its localization in vivo can be determined by fluorescence microscopy. The target gene should be cloned into pDsRed-Express-C1 so that it is in frame with the DsRed-Express coding sequences, with no intervening in-frame stop codons. The recombinant DsRed-Express vector can be transfected into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (5). pDsRed-Express-C1 can also be used as a cotransfection marker; the unmodified vector will express DsRed-Express.
pDsRed-Express-C1载体含有以下元件:
Human cytomegalovirus (CMV) immediate early promoter: 1–589
Enhancer region: 59–465;
TATA box: 554–560
Transcription start point: 583
C→G mutation to remove Sac I site: 569
Discosoma sp. human codon-optimized Red Fluorescent Protein (DsRed-Express) gene
Kozak consensus translation initiation site: 606–616
Start codon (ATG): 613–615; Stop codon: 1357–1359
CGC→GCC (Arg-2 to Ala) mutation: 616–618
AAG→GAG (Lys-5 to Glu) mutation: 625–627
AAC→GAC (Asn-6 to Asp) mutation: 628–630
ACC→TCC (Thr-21 to Ser) mutation: 673–675
CAC→ACC (His-41 to Thr) mutation: 733–735
AAC→CAG (Asn-42 to Gln) mutation: 736–738
GTG→GCC (Val-44 to Ala) mutation: 742–744
TGC→TCC (Cys-117 to Ser) mutation: 961–963
ACC→GCC (Thr-217 to Ala) mutation: 1261–1263
Last amino acid in DsRed-Express: 1285–1287
MCS: 1288–1344
SV40 early mRNA polyadenylation signal
Polyadenylation signals: 1498–1503 & 1527–1532; mRNA
3' ends: 1536 & 1648
f1 single-strand DNA origin: 1595–2050 (Packages the noncoding strand of DsRed-Express.)
Bacterial promoter for expression of Kanr gene
–35 region: 21
12–2117; –10 region: 2135–2140
Transcription start point: 2147
SV40 origin of replication: 2391–2526
SV40 early promoter
Enhancer (72-bp tandem repeats): 2224–2295 & 2296–2367
21-bp repeats: 2371–2991, 2392–2412 & 2414–2431
Early promoter element: 2447–2453
Major transcription start points: 2443, 2481, 2487 & 2492
Kanamycin/neomycin resistance gene
Neomycin phosphotransferase coding sequences:
Start codon (ATG): 2576–2578; stop codon: 3368–3370
G→A mutation to remove Pst I site: 2758
C→A (Arg to Ser) mutation to remove BssH II site: 3104
Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal
Polyadenylation signals: 3606–3611 & 3619–3624
pUC plasmid replication origin: 3955–4598
Propagation in E. coli
Suitable host strains: DH5α, HB101, and other general purpose strains. Single-stranded DNA production
requires a host containing an F plasmid such as JM109 or XL1-Blue.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) to E. coli hosts.
E. coli replication origin: pUC
Copy number: ≈500
Plasmid incompatibility group: pMB1/Col E1
Excitation and emission maxima of DsRed-Express
Excitation maximum = 557 nm
Emission maximum = 579 nm
载体序列
ORIGIN
1 TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA TGGAGTTCCG
61 CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT
121 GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA
181 ATGGGTGGAG TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC
241 AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA
301 CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC
361 CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG
421 ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG
481 GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT
541 ACGGTGGGAG GTCTATATAA GCAGAGCTGG TTTAGTGAAC CGTCAGATCC GCTAGCGCTA
601 CCGGTCGCCA CCATGGCCTC CTCCGAGGAC GTCATCAAGG AGTTCATGCG CTTCAAGGTG
661 CGCATGGAGG GCTCCGTGAA CGGCCACGAG TTCGAGATCG AGGGCGAGGG CGAGGGCCGC
721 CCCTACGAGG GCACCCAGAC CGCCAAGCTG AAGGTGACCA AGGGCGGCCC CCTGCCCTTC
781 GCCTGGGACA TCCTGTCCCC CCAGTTCCAG TACGGCTCCA AGGTGTACGT GAAGCACCCC
841 GCCGACATCC CCGACTACAA GAAGCTGTCC TTCCCCGAGG GCTTCAAGTG GGAGCGCGTG
901 ATGAACTTCG AGGACGGCGG CGTGGTGACC GTGACCCAGG ACTCCTCCCT GCAGGACGGC
961 TCCTTCATCT ACAAGGTGAA GTTCATCGGC GTGAACTTCC CCTCCGACGG CCCCGTAATG
1021 CAGAAGAAGA CTATGGGCTG GGAGGCCTCC ACCGAGCGCC TGTACCCCCG CGACGGCGTG
1081 CTGAAGGGCG AGATCCACAA GGCCCTGAAG CTGAAGGACG GCGGCCACTA CCTGGTGGAG
1141 TTCAAGTCCA TCTACATGGC CAAGAAGCCC GTGCAGCTGC CCGGCTACTA CTACGTGGAC
1201 TCCAAGCTGG ACATCACCTC CCACAACGAG GACTACACCA TCGTGGAGCA GTACGAGCGC
1261 GCCGAGGGCC GCCACCACCT GTTCCTGAGA TCTCGAGCTC AAGCTTCGAA TTCTGCAGTC
1321 GACGGTACCG CGGGCCCGGG ATCCACCGGA TCTAGATAAC TGATCATAAT CAGCCATACC
1381 ACATTTGTAG AGGTTTTACT TGCTTTAAAA AACCTCCCAC ACCTCCCCCT GAACCTGAAA
1441 CATAAAATGA ATGCAATTGT TGTTGTTAAC TTGTTTATTG CAGCTTATAA TGGTTACAAA
1501 TAAAGCAATA GCATCACAAA TTTCACAAAT AAAGCATTTT TTTCACTGCA TTCTAGTTGT
1561 GGTTTGTCCA AACTCATCAA TGTATCTTAA CGCGTAAATT GTAAGCGTTA ATATTTTGTT
1621 AAAATTCGCG TTAAATTTTT GTTAAATCAG CTCATTTTTT AACCAATAGG CCGAAATCGG
1681 CAAAATCCCT TATAAATCAA AAGAATAGAC CGAGATAGGG TTGAGTGTTG TTCCAGTTTG
1741 GAACAAGAGT CCACTATTAA AGAACGTGGA CTCCAACGTC AAAGGGCGAA AAACCGTCTA
1801 TCAGGGCGAT GGCCCACTAC GTGAACCATC ACCCTAATCA AGTTTTTTGG GGTCGAGGTG
1861 CCGTAAAGCA CTAAATCGGA ACCCTAAAGG GAGCCCCCGA TTTAGAGCTT GACGGGGAAA
1921 GCCGGCGAAC GTGGCGAGAA AGGAAGGGAA GAAAGCGAAA GGAGCGGGCG CTAGGGCGCT
1981 GGCAAGTGTA GCGGTCACGC TGCGCGTAAC CACCACACCC GCCGCGCTTA ATGCGCCGCT
2041 ACAGGGCGCG TCAGGTGGCA CTTTTCGGGG AAATGTGCGC GGAACCCCTA TTTGTTTATT
2101 TTTCTAAATA CATTCAAATA TGTATCCGCT CATGAGACAA TAACCCTGAT AAATGCTTCA
2161 ATAATATTGA AAAAGGAAGA GTCCTGAGGC GGAAAGAACC AGCTGTGGAA TGTGTGTCAG
2221 TTAGGGTGTG GAAAGTCCCC AGGCTCCCCA GCAGGCAGAA GTATGCAAAG CATGCATCTC
2281 AATTAGTCAG CAACCAGGTG TGGAAAGTCC CCAGGCTCCC CAGCAGGCAG AAGTATGCAA
2341 AGCATGCATC TCAATTAGTC AGCAACCATA GTCCCGCCCC TAACTCCGCC CATCCCGCCC
2401 CTAACTCCGC CCAGTTCCGC CCATTCTCCG CCCCATGGCT GACTAATTTT TTTTATTTAT
2461 GCAGAGGCCG AGGCCGCCTC GGCCTCTGAG CTATTCCAGA AGTAGTGAGG AGGCTTTTTT
2521 GGAGGCCTAG GCTTTTGCAA AGATCGATCA AGAGACAGGA TGAGGATCGT TTCGCATGAT
2581 TGAACAAGAT GGATTGCACG CAGGTTCTCC GGCCGCTTGG GTGGAGAGGC TATTCGGCTA
2641 TGACTGGGCA CAACAGACAA TCGGCTGCTC TGATGCCGCC GTGTTCCGGC TGTCAGCGCA
2701 GGGGCGCCCG GTTCTTTTTG TCAAGACCGA CCTGTCCGGT GCCCTGAATG AACTGCAAGA
2761 CGAGGCAGCG CGGCTATCGT GGCTGGCCAC GACGGGCGTT CCTTGCGCAG CTGTGCTCGA
2821 CGTTGTCACT GAAGCGGGAA GGGACTGGCT GCTATTGGGC GAAGTGCCGG GGCAGGATCT
2881 CCTGTCATCT CACCTTGCTC CTGCCGAGAA AGTATCCATC ATGGCTGATG CAATGCGGCG
2941 GCTGCATACG CTTGATCCGG CTACCTGCCC ATTCGACCAC CAAGCGAAAC ATCGCATCGA
3001 GCGAGCACGT ACTCGGATGG AAGCCGGTCT TGTCGATCAG GATGATCTGG ACGAAGAGCA
3061 TCAGGGGCTC GCGCCAGCCG AACTGTTCGC CAGGCTCAAG GCGAGCATGC CCGACGGCGA
3121 GGATCTCGTC GTGACCCATG GCGATGCCTG CTTGCCGAAT ATCATGGTGG AAAATGGCCG
3181 CTTTTCTGGA TTCATCGACT GTGGCCGGCT GGGTGTGGCG GACCGCTATC AGGACATAGC
3241 GTTGGCTACC CGTGATATTG CTGAAGAGCT TGGCGGCGAA TGGGCTGACC GCTTCCTCGT
3301 GCTTTACGGT ATCGCCGCTC CCGATTCGCA GCGCATCGCC TTCTATCGCC TTCTTGACGA
3361 GTTCTTCTGA GCGGGACTCT GGGGTTCGAA ATGACCGACC AAGCGACGCC CAACCTGCCA
3421 TCACGAGATT TCGATTCCAC CGCCGCCTTC TATGAAAGGT TGGGCTTCGG AATCGTTTTC
3481 CGGGACGCCG GCTGGATGAT CCTCCAGCGC GGGGATCTCA TGCTGGAGTT CTTCGCCCAC
3541 CCTAGGGGGA GGCTAACTGA AACACGGAAG GAGACAATAC CGGAAGGAAC CCGCGCTATG
3601 ACGGCAATAA AAAGACAGAA TAAAACGCAC GGTGTTGGGT CGTTTGTTCA TAAACGCGGG
3661 GTTCGGTCCC AGGGCTGGCA CTCTGTCGAT ACCCCACCGA GACCCCATTG GGGCCAATAC
3721 GCCCGCGTTT CTTCCTTTTC CCCACCCCAC CCCCCAAGTT CGGGTGAAGG CCCAGGGCTC
3781 GCAGCCAACG TCGGGGCGGC AGGCCCTGCC ATAGCCTCAG GTTACTCATA TATACTTTAG
3841 ATTGATTTAA AACTTCATTT TTAATTTAAA AGGATCTAGG TGAAGATCCT TTTTGATAAT
3901 CTCATGACCA AAATCCCTTA ACGTGAGTTT TCGTTCCACT GAGCGTCAGA CCCCGTAGAA
3961 AAGATCAAAG GATCTTCTTG AGATCCTTTT TTTCTGCGCG TAATCTGCTG CTTGCAAACA
4021 AAAAAACCAC CGCTACCAGC GGTGGTTTGT TTGCCGGATC AAGAGCTACC AACTCTTTTT
4081 CCGAAGGTAA CTGGCTTCAG CAGAGCGCAG ATACCAAATA CTGTCCTTCT AGTGTAGCCG
4141 TAGTTAGGCC ACCACTTCAA GAACTCTGTA GCACCGCCTA CATACCTCGC TCTGCTAATC
4201 CTGTTACCAG TGGCTGCTGC CAGTGGCGAT AAGTCGTGTC TTACCGGGTT GGACTCAAGA
4261 CGATAGTTAC CGGATAAGGC GCAGCGGTCG GGCTGAACGG GGGGTTCGTG CACACAGCCC
4321 AGCTTGGAGC GAACGACCTA CACCGAACTG AGATACCTAC AGCGTGAGCT ATGAGAAAGC
4381 GCCACGCTTC CCGAAGGGAG AAAGGCGGAC AGGTATCCGG TAAGCGGCAG GGTCGGAACA
4441 GGAGAGCGCA CGAGGGAGCT TCCAGGGGGA AACGCCTGGT ATCTTTATAG TCCTGTCGGG
4501 TTTCGCCACC TCTGACTTGA GCGTCGATTT TTGTGATGCT CGTCAGGGGG GCGGAGCCTA
4561 TGGAAAAACG CCAGCAACGC GGCCTTTTTA CGGTTCCTGG CCTTTTGCTG GCCTTTTGCT
4621 CACATGTTCT TTCCTGCGTT ATCCCCTGAT TCTGTGGATA ACCGTATTAC CGCCATGCAT
//
其他哺乳动物表达载体:
pcDNA6.2/nGeneBLAzer-GW/D-TOPO |


