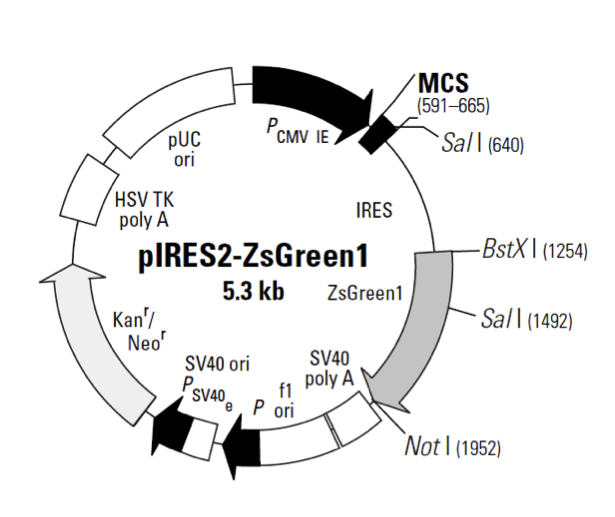
plRES2-ZsGreen1
plRES2-ZsGreen1
编号 | 载体名称 |
北京华越洋生物VECT6130 | plRES2-ZsGreen1 |
plRES2-ZsGreen1载体基本信息
载体名称: | plRES2-ZsGreen1 |
质粒类型: | 哺乳动物表达载体 ; 双顺反子载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 多克隆位点,限制性内切酶 |
启动子: | CMV |
载体大小: | 5.3kb |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | 无 |
载体抗性: | 卡那霉素 |
筛选标记: | -- |
备注: | ZsGreen1比EGFP亮度更高; |
稳定性: | -- |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
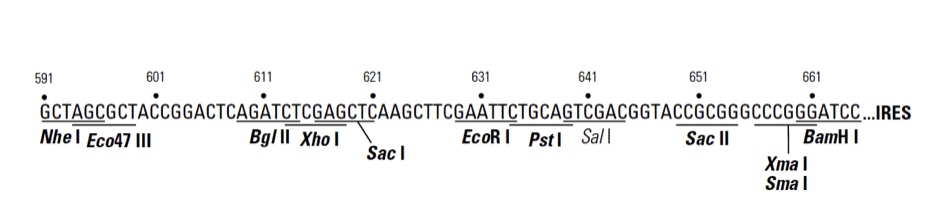
plRES2-ZsGreen1载体质粒图谱和多克隆位点信息

plRES2-ZsGreen1载体简介
IRES-containing bicistronic vectors allow the simultaneous expression of two proteins separately but from the same RNA transcript (1, 2).
How Does an IRES Work?
The IRES of the encephalomyocarditis virus (ECMV) permits the translation of two open reading frames from one messenger RNA. Although translation initiation of eukaryotic mRNAs occurs almost exclusively at the 5' cap, the IRES allows ribosomes to bind and initiate translation at a second, internal location. Thus, two proteins are expressed simultaneously from the same bicistronic mRNA transcript.Clontech's pIRES Vector contains two multiple cloning sites flanking the central IRES for you to clone and express two genes of interest.pIRES Vectors Containing Antibiotic Selection Markers
The pIRES Selection Vectors pIRESpuro3, pIREShyg3, pIRESneo3, and pIRESbleo3 each contain a multiple cloning site located upstream of the IRES from which a selection marker is expressed. Because your gene of interest and a selection marker are translated from a single RNA, you can be certain that nearly 100% of the colonies that are resistant to puromycin, hygromycin, G418, and bleomycin also express your protein of interest.
All IRES Selection Vectors contain a partially disabled IRES which reduces the efficiency of translation initiation for the selection marker relative to that of the cloned gene, and allows preferential selection of cells expressing high levels of your protein of interest (3).
Bicistronic (IRES) Fluorescent Protein Expression Vectors
Similarly, fluorescent protein-containing IRES vectors allow coexpression of your target protein and a fluorescent protein.
Successfully transfected target cells are easily identified by fluorescence microscopy or flow cytometry. The are several options for different fluorescent proteins, which include mCherry, ZsGreen1, and tdTomato (pLVX-IRES Vectors) and AcGFP1, DsRed2, and DsRed-Express (pIRES2 Vectors).
Features
Translate your gene of interest and an antibiotic resistance marker or fluorescent protein from a single mRNA
Quickly identify cells expressing your protein of interest at high levels
Bicistronic expression allows faster, better stable clone selection
Choice of vectors enables screening via antibiotic selection, fluorescence microscopy, or flow cytometry
Applications
Rapidly select stable clones that show high-level expression of your target protein
Identify transfected cells by antibiotic selection, fluorescence microscopy, or flow cytometry
References
Jackson, R. J. et al. (1990) Trends Biochem. Sci. 15:477–483.
Jang, S. K. et al. (1988) J. Virol. 62:2636–2643.
Rees, S. et al. (1996) BioTechniques 20:102–110.
plRES2-ZsGreen1其他哺乳动物表达载体: