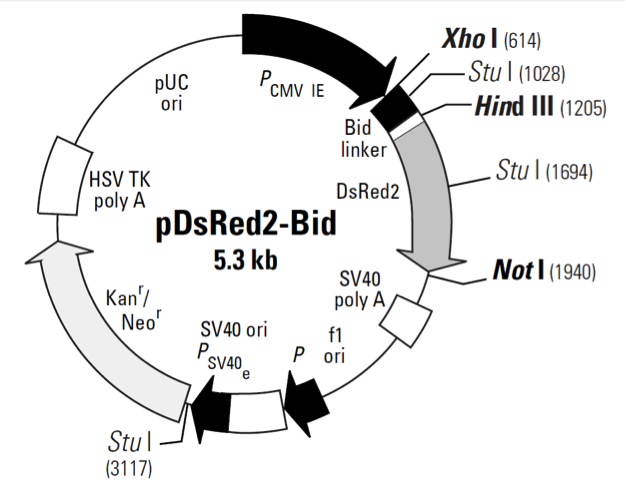
pDsRed2-Bid
pDsRed2-Bid
编号 | 载体名称 |
北京华越洋生物VECT6108 | pDsRed2-Bid |
pDsRed2-Bid载体基本信息
载体名称: | pDsRed2-Bid |
质粒类型: | 哺乳动物细胞表达载体;信号通路报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | CMV IE |
载体大小: | 5.3 kb |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | -- |
载体抗性: | 卡那霉素 |
筛选标记: | 新霉素(Neomycin) |
克隆菌株: | DH5α, HB101 |
宿主细胞(系): | 常规细胞系,293、CV-1、CHO等 |
备注: | pDsRed2-Bid载体是红色荧光报告载体,用于研究依赖Bid的细胞凋亡信号通路。 |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
pDsRed2-Bid载体质粒图谱和多克隆位点信息
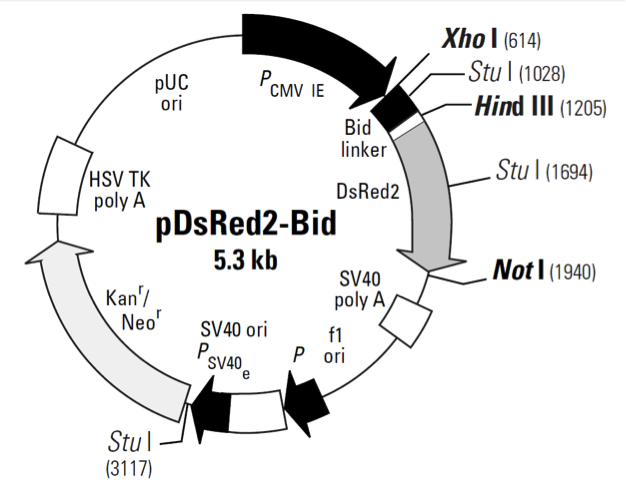

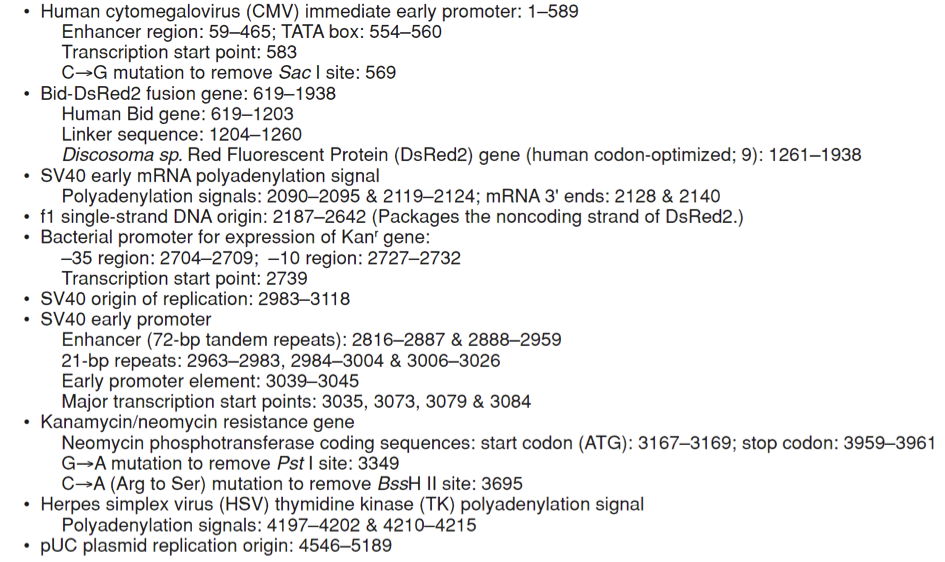
pDsRed2-Bid载体描述
pDsRed2-Bid is a mammalian expression vector that encodes a fusion of Discosoma sp. red fluorescent protein (DsRed2; 1, 2) and Bid, a member of the Bcl-2 “pro-apoptosis” family (3). Developed for our ApoAlert line, pDsRed2-Bid is designed to help researchers study Bid-dependent apoptosis pathways. Because of its fluorescent label, the Bid-DsRed2 fusion is easily detected by microscopy, allowing researchers to track its movements in response to certain apoptosis inducing agents (e.g., TNF-α).
Bid’s activity is closely tied to its location in the cell. In healthy, non-apoptotic cells, Bid normally resides in the cytosol. But soon after the induction of apoptosis, it translocates to mitochondria, where it stimulates the release of cytochrome c—a key amplification step in the apoptotic cascade (4–7). The translocation is triggered by caspase-8, which, when activated by a death signal, cleaves Bid to produce a 15-kDa C-terminal fragment. The fragment, often referred to as truncated Bid or tBid, transmits the death signal further by translocating to the mitochondria. You can monitor this event visually by expressing Bid as a DsRed2 fusion. Bid-DsRed2, the fusion expressed by this vector, for example, emits red fluorescence—even after truncation—so it can be followed as it moves from the cytosol to the mitochondria. To drive expression of Bid-DsRed2, this vector contains the immediate early promoter of cytomegalovirus (PCMV IE), positioned just upstream of the Bid sequence. A short linker joins the Bid coding sequence to the 5'-end of DsRed2. Farther downstream, the vector contains a pair of SV40 polyadenylation signals, which direct proper processing of the 3'-end of the Bid-DsRed2 mRNA. The vector also contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor), consisting of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene, allows stably transfected eukaryotic cells to be selected using G418 (8). A bacterial promoter upstream of the cassette confers kanamycin resistance (Kanr) to E. coli.
Cells transfected with pDsRed2-Bid constitutively express Bid as an N-terminal fusion to DsRed2. pDsRed2-Bid can be introduced into a variety of mammalian cell lines using any standard transfection method. If required, stable transfectants can be selected using G418 (8).
Propagation in E. coli
Suitable host strains: DH5α, HB101 and other general purpose strains. Single-stranded DNA production requires
a host containing an F plasmid such as JM109 or XL1-Blue.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) to E. coli hosts.
E. coli replication origin: pUC
Copy number: ~500
Plasmid incompatibility group: pMB1/ColE1
Red Fluorescent Protein (DsRed2)
Excitation/Emission Maxima: 558 nm / 583 nm
其他哺乳动物表达载体: