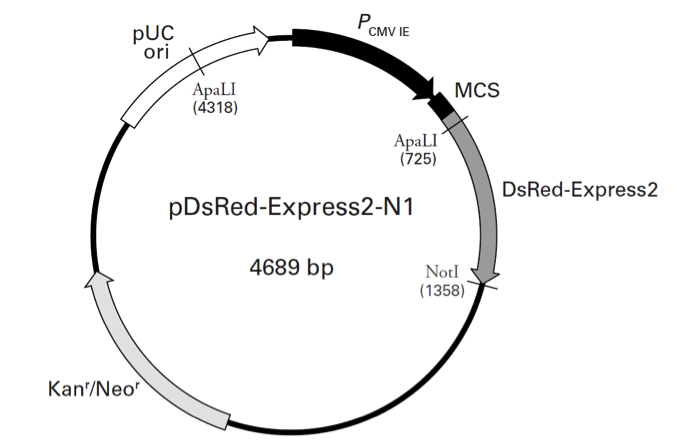
pDsRed-Express2-N1
pDsRed-Express2-N1
编号 | 载体名称 |
北京华越洋生物VECT6161 | pDsRed-Express2-N1 |
pDsRed-Express2-N1载体基本信息
载体名称: | pDsRed-Express2-N1 |
质粒类型: | 哺乳动物细胞表达载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | CMV IE |
载体大小: | 4689 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | DsRed-Express2(C-端) |
载体抗性: | 卡那霉素 |
筛选标记: | 新霉素(Neomycin) |
克隆菌株: | DH5α, HB101 |
宿主细胞(系): | 哺乳动物细胞系(包括原代细胞和干细胞) |
备注: | pDsRed-Express2-N1载体是荧光报告载体,表达N端DsRed-Express2融合蛋白; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
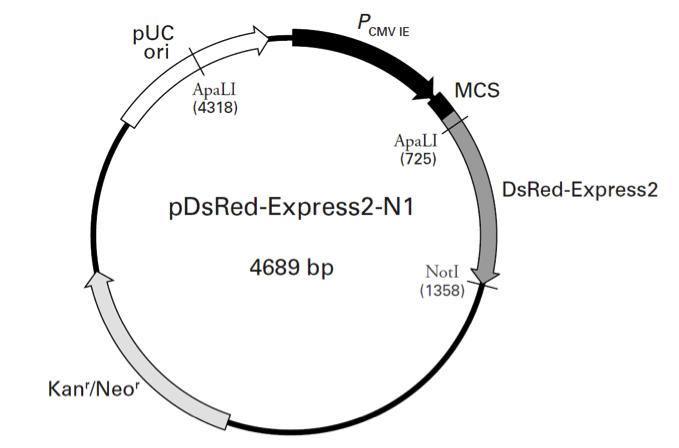
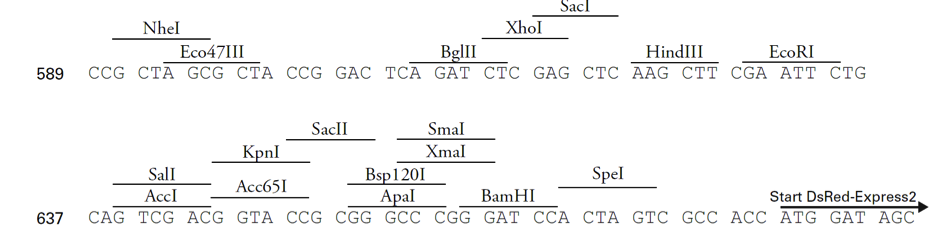
pDsRed-Express2-N1载体质粒图谱和多克隆位点信息
pDsRed-Express2-N1载体描述
pDsRed-Express2-N1 is a mammalian expression vector designed to express a protein of interest fused to the N-terminus of DsRed-Express2, a variant of the Discosoma sp. red fluorescent protein, DsRed (1). DsRed-Express2 retains the fast maturation and high photostability characteristic of its predecessor, DsRed-Express (2), and has been engineered (through additional amino acid substitutions) for increased solubility (3). Although it most likely forms the same tetrameric structure as wild-type DsRed, DsRed-Express2 displays a greatly reduced tendency to aggregate, resulting in reduced cyto- and phototoxicity, and making DsRed-Express2 much better suited for in vivo applications involving sensitive cells, such as primary or stem cells. (Please note: Because DsRed-Express2 likely forms tetramers, its suitability as a fusion partner will largely depend on how its tetramerization affects the function of the protein to which it is fused.) DsRed-Express2 also exhibits extremely low residual green fluorescence, which allows cells expressing the protein to be effectively separated from other fluorescently labeled cell populations by flow cytometry. The multiple cloning site (MCS) in pDsRed-Express2-N1 is positioned upstream of the DsRed-Express2 coding sequence. A Kozak consensus sequence (4), located between the MCS and the DsRed-Express2 coding sequence, enhances the translational efficiency of the unfused DsRed-Express2 protein in eukaryotic cells. SV40 polyadenylation signals downstream of the DsRed-Express2 coding sequence direct proper processing of the 3' ends of the DsRed-Express2 and fusion gene mRNA transcripts.
The vector backbone also contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. This vector also has a neomycin-resistance cassette (Neor) that allows G418 selection of stably transfected eukaryotic cells (5). This cassette consists of the SV40 early promoter, a Tn5 kanamycin/neomycin resistance gene, and herpes simplex virus thymidine kinase (HSV TK) polyadenylation signals . A bacterial promoter upstream of this cassette allows kanamycin resistance in E. coli.
To construct a fusion protein, the gene of interest must be cloned into pDsRed-Express2-N1 so that it is in-frame with the DsRed-Express2 coding sequence; the gene must include an initiation codon (ATG), and lack in-frame stop codons. pDsRed-Express2-N1 can also be used as a cotransfection marker, as the unmodified vector will express DsRed-Express2 in mammalian cells.
pDsRed-Express2-N1 can be transfected into mammalian cells using any standard transfection method. Fusions that retain the fluorescence properties of the native DsRed-Express2 protein (excitation and emission maxima: 541 and 591, respectively) can be monitored by flow cytometry and localized by fluorescence microscopy. If required, stable transfectants can be selected using G418.
For Western analysis, DsRed-Express2 can be detected with either the Living Colors DsRed Polyclonal Anti-body (Cat. No. 632496) or the Living Colors DsRed Monoclonal Antibody (Cat. Nos. 632392 and 632393).
pDsRed-Express2-N1载体含有以下元件:
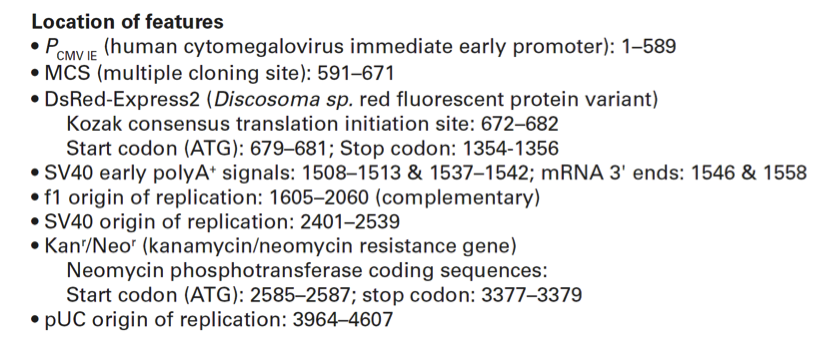
PCMV IE (human cytomegalovirus immediate early promoter): 1–589
MCS (multiple cloning site): 591–671
DsRed-Express2 (Discosoma sp. red fluorescent protein variant)
Kozak consensus translation initiation site: 672–682
Start codon (ATG): 679–681; Stop codon: 1354-1356
SV40 early polyA+ signals: 1508–1513 & 1537–1542; mRNA 3' ends: 1546 & 1558
f1 origin of replication: 1605–2060 (complementary)
SV40 origin of replication: 2401–2539
Kanr/Neor (kanamycin/neomycin resistance gene)
Neomycin phosphotransferase coding sequences:
Start codon (ATG): 2585–2587; stop codon: 3377–3379
pUC origin of replication: 3964–4607
Propagation in E. coli
Recommended host strain: DH5α, HB101, and other general purpose strains.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts.
E. coli replication origin: pUC
Copy number: high
Plasmid incompatibility group: pMB1/ColE1
Excitation and emission maxima of DsRed-Express2
Excitation maximum = 554 nm
Emission maximum = 591 nm
pDsRed-Express2-N1载体序列
ORIGIN
1 TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA TGGAGTTCCG
61 CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT
121 GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA
181 ATGGGTGGAG TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC
241 AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA
301 CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC
361 CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG
421 ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG
481 GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT
541 ACGGTGGGAG GTCTATATAA GCAGAGCTGG TTTAGTGAAC CGTCAGATCC GCTAGCGCTA
601 CCGGACTCAG ATCTCGAGCT CAAGCTTCGA ATTCTGCAGT CGACGGTACC GCGGGCCCGG
661 GATCCACTAG TCGCCACCAT GGATAGCACT GAGAACGTCA TCAAGCCCTT CATGCGCTTC
721 AAGGTGCACA TGGAGGGCTC CGTGAACGGC CACGAGTTCG AGATCGAGGG CGAGGGCGAG
781 GGCAAGCCCT ACGAGGGCAC CCAGACCGCC AAGCTGCAGG TGACCAAGGG CGGCCCCCTG
841 CCCTTCGCCT GGGACATCCT GTCCCCCCAG TTCCAGTACG GCTCCAAGGT GTACGTGAAG
901 CACCCCGCCG ACATCCCCGA CTACAAGAAG CTGTCCTTCC CCGAGGGCTT CAAGTGGGAG
961 CGCGTGATGA ACTTCGAGGA CGGCGGCGTG GTGACCGTGA CCCAGGACTC CTCCCTGCAG
1021 GACGGCACCT TCATCTACCA CGTGAAGTTC ATCGGCGTGA ACTTCCCCTC CGACGGCCCC
1081 GTAATGCAGA AGAAGACTCT GGGCTGGGAG CCCTCCACCG AGCGCCTGTA CCCCCGCGAC
1141 GGCGTGCTGA AGGGCGAGAT CCACAAGGCG CTGAAGCTGA AGGGCGGCGG CCACTACCTG
1201 GTGGAGTTCA AGTCAATCTA CATGGCCAAG AAGCCCGTGA AGCTGCCCGG CTACTACTAC
1261 GTGGACTCCA AGCTGGACAT CACCTCCCAC AACGAGGACT ACACCGTGGT GGAGCAGTAC
1321 GAGCGCGCCG AGGCCCGCCA CCACCTGTTC CAGTAGCGGC CGCGACTCTA GATCATAATC
1381 AGCCATACCA CATTTGTAGA GGTTTTACTT GCTTTAAAAA ACCTCCCACA CCTCCCCCTG
1441 AACCTGAAAC ATAAAATGAA TGCAATTGTT GTTGTTAACT TGTTTATTGC AGCTTATAAT
1501 GGTTACAAAT AAAGCAATAG CATCACAAAT TTCACAAATA AAGCATTTTT TTCACTGCAT
1561 TCTAGTTGTG GTTTGTCCAA ACTCATCAAT GTATCTTAAG GCGTAAATTG TAAGCGTTAA
1621 TATTTTGTTA AAATTCGCGT TAAATTTTTG TTAAATCAGC TCATTTTTTA ACCAATAGGC
1681 CGAAATCGGC AAAATCCCTT ATAAATCAAA AGAATAGACC GAGATAGGGT TGAGTGTTGT
1741 TCCAGTTTGG AACAAGAGTC CACTATTAAA GAACGTGGAC TCCAACGTCA AAGGGCGAAA
1801 AACCGTCTAT CAGGGCGATG GCCCACTACG TGAACCATCA CCCTAATCAA GTTTTTTGGG
1861 GTCGAGGTGC CGTAAAGCAC TAAATCGGAA CCCTAAAGGG AGCCCCCGAT TTAGAGCTTG
1921 ACGGGGAAAG CCGGCGAACG TGGCGAGAAA GGAAGGGAAG AAAGCGAAAG GAGCGGGCGC
1981 TAGGGCGCTG GCAAGTGTAG CGGTCACGCT GCGCGTAACC ACCACACCCG CCGCGCTTAA
2041 TGCGCCGCTA CAGGGCGCGT CAGGTGGCAC TTTTCGGGGA AATGTGCGCG GAACCCCTAT
2101 TTGTTTATTT TTCTAAATAC ATTCAAATAT GTATCCGCTC ATGAGACAAT AACCCTGATA
2161 AATGCTTCAA TAATATTGAA AAAGGAAGAG TCCTGAGGCG GAAAGAACCA GCTGTGGAAT
2221 GTGTGTCAGT TAGGGTGTGG AAAGTCCCCA GGCTCCCCAG CAGGCAGAAG TATGCAAAGC
2281 ATGCATCTCA ATTAGTCAGC AACCAGGTGT GGAAAGTCCC CAGGCTCCCC AGCAGGCAGA
2341 AGTATGCAAA GCATGCATCT CAATTAGTCA GCAACCATAG TCCCGCCCCT AACTCCGCCC
2401 ATCCCGCCCC TAACTCCGCC CAGTTCCGCC CATTCTCCGC CCCATGGCTG ACTAATTTTT
2461 TTTATTTATG CAGAGGCCGA GGCCGCCTCG GCCTCTGAGC TATTCCAGAA GTAGTGAGGA
2521 GGCTTTTTTG GAGGCCTAGG CTTTTGCAAA GATCGATCAA GAGACAGGAT GAGGATCGTT
2581 TCGCATGATT GAACAAGATG GATTGCACGC AGGTTCTCCG GCCGCTTGGG TGGAGAGGCT
2641 ATTCGGCTAT GACTGGGCAC AACAGACAAT CGGCTGCTCT GATGCCGCCG TGTTCCGGCT
2701 GTCAGCGCAG GGGCGCCCGG TTCTTTTTGT CAAGACCGAC CTGTCCGGTG CCCTGAATGA
2761 ACTGCAAGAC GAGGCAGCGC GGCTATCGTG GCTGGCCACG ACGGGCGTTC CTTGCGCAGC
2821 TGTGCTCGAC GTTGTCACTG AAGCGGGAAG GGACTGGCTG CTATTGGGCG AAGTGCCGGG
2881 GCAGGATCTC CTGTCATCTC ACCTTGCTCC TGCCGAGAAA GTATCCATCA TGGCTGATGC
2941 AATGCGGCGG CTGCATACGC TTGATCCGGC TACCTGCCCA TTCGACCACC AAGCGAAACA
3001 TCGCATCGAG CGAGCACGTA CTCGGATGGA AGCCGGTCTT GTCGATCAGG ATGATCTGGA
3061 CGAAGAGCAT CAGGGGCTCG CGCCAGCCGA ACTGTTCGCC AGGCTCAAGG CGAGCATGCC
3121 CGACGGCGAG GATCTCGTCG TGACCCATGG CGATGCCTGC TTGCCGAATA TCATGGTGGA
3181 AAATGGCCGC TTTTCTGGAT TCATCGACTG TGGCCGGCTG GGTGTGGCGG ACCGCTATCA
3241 GGACATAGCG TTGGCTACCC GTGATATTGC TGAAGAGCTT GGCGGCGAAT GGGCTGACCG
3301 CTTCCTCGTG CTTTACGGTA TCGCCGCTCC CGATTCGCAG CGCATCGCCT TCTATCGCCT
3361 TCTTGACGAG TTCTTCTGAG CGGGACTCTG GGGTTCGAAA TGACCGACCA AGCGACGCCC
3421 AACCTGCCAT CACGAGATTT CGATTCCACC GCCGCCTTCT ATGAAAGGTT GGGCTTCGGA
3481 ATCGTTTTCC GGGACGCCGG CTGGATGATC CTCCAGCGCG GGGATCTCAT GCTGGAGTTC
3541 TTCGCCCACC CTAGGGGGAG GCTAACTGAA ACACGGAAGG AGACAATACC GGAAGGAACC
3601 CGCGCTATGA CGGCAATAAA AAGACAGAAT AAAACGCACG GTGTTGGGTC GTTTGTTCAT
3661 AAACGCGGGG TTCGGTCCCA GGGCTGGCAC TCTGTCGATA CCCCACCGAG ACCCCATTGG
3721 GGCCAATACG CCCGCGTTTC TTCCTTTTCC CCACCCCACC CCCCAAGTTC GGGTGAAGGC
3781 CCAGGGCTCG CAGCCAACGT CGGGGCGGCA GGCCCTGCCA TAGCCTCAGG TTACTCATAT
3841 ATACTTTAGA TTGATTTAAA ACTTCATTTT TAATTTAAAA GGATCTAGGT GAAGATCCTT
3901 TTTGATAATC TCATGACCAA AATCCCTTAA CGTGAGTTTT CGTTCCACTG AGCGTCAGAC
3961 CCCGTAGAAA AGATCAAAGG ATCTTCTTGA GATCCTTTTT TTCTGCGCGT AATCTGCTGC
4021 TTGCAAACAA AAAAACCACC GCTACCAGCG GTGGTTTGTT TGCCGGATCA AGAGCTACCA
4081 ACTCTTTTTC CGAAGGTAAC TGGCTTCAGC AGAGCGCAGA TACCAAATAC TGTTCTTCTA
4141 GTGTAGCCGT AGTTAGGCCA CCACTTCAAG AACTCTGTAG CACCGCCTAC ATACCTCGCT
4201 CTGCTAATCC TGTTACCAGT GGCTGCTGCC AGTGGCGATA AGTCGTGTCT TACCGGGTTG
4261 GACTCAAGAC GATAGTTACC GGATAAGGCG CAGCGGTCGG GCTGAACGGG GGGTTCGTGC
4321 ACACAGCCCA GCTTGGAGCG AACGACCTAC ACCGAACTGA GATACCTACA GCGTGAGCTA
4381 TGAGAAAGCG CCACGCTTCC CGAAGGGAGA AAGGCGGACA GGTATCCGGT AAGCGGCAGG
4441 GTCGGAACAG GAGAGCGCAC GAGGGAGCTT CCAGGGGGAA ACGCCTGGTA TCTTTATAGT
4501 CCTGTCGGGT TTCGCCACCT CTGACTTGAG CGTCGATTTT TGTGATGCTC GTCAGGGGGG
4561 CGGAGCCTAT GGAAAAACGC CAGCAACGCG GCCTTTTTAC GGTTCCTGGC CTTTTGCTGG
4621 CCTTTTGCTC ACATGTTCTT TCCTGCGTTA TCCCCTGATT CTGTGGATAA CCGTATTACC
4681 GCCATGCAT
//
其他哺乳动物表达载体: