pEF1α-AcGFP1-N1
pEF1α-AcGFP1-N1
编号 | 载体名称 |
北京华越洋生物VECT6134 | pEF1α-AcGFP1-N1 |
pEF1α-AcGFP1-N1载体基本信息
载体名称: | pEF1α-AcGFP1-N1 |
质粒类型: | 哺乳动物表达载体;双顺反子载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 多克隆位点,限制性内切酶 |
启动子: | EF1α |
载体大小: | 5483 bp |
5' 测序引物及序列: | EF1-F: TCAAGCCTCAGACAGTGGTTC |
3' 测序引物及序列: | IRES-R |
载体标签: | 无标签 |
载体抗性: | 卡那霉素 |
筛选标记: | Neomycin、AcGFP |
克隆菌株: | DH5alpha |
宿主细胞(系): | 大部分细胞类型,包括原代细胞、分化细胞、干细胞。 |
备注: | pEF1α-AcGFP1-N1载体是哺乳动物表达载体; |
稳定性: | 瞬表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
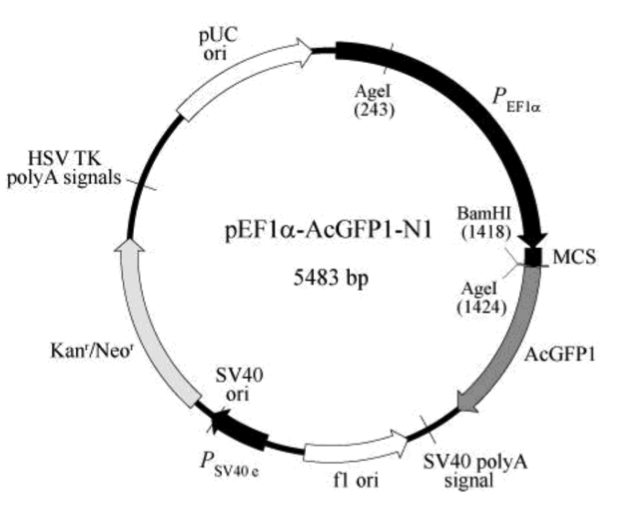
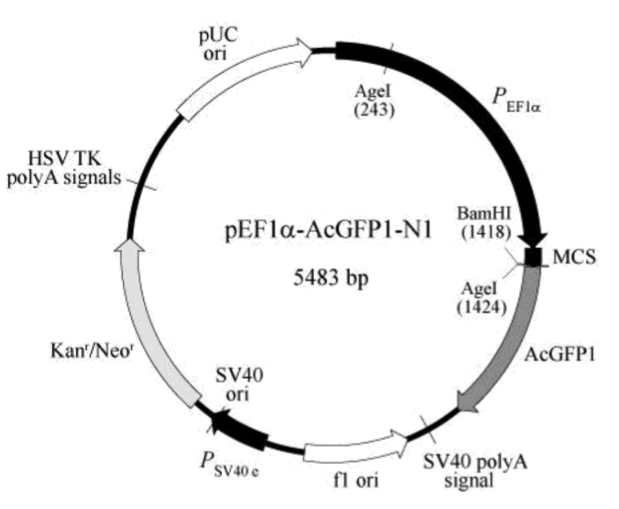
pEF1α-AcGFP1-N1载体质粒图谱和多克隆位点信息

pEF1α-AcGFP1-N1载体简介
pEF1α-AcGFP1-N1 is designed to express a protein of interest fused to the N-terminus of AcGFP1, a human-codonoptimized, monomeric green fluorescent protein derived from Aequorea coerulescens. The excitation and emission maxima of the native AcGFP1 protein are 475 nm and 505 nm, respectively. Expression of fusion proteins that retain the fluorescence properties of the unmodified AcGFP1 protein can be monitored by flow cytometry and localized by fluorescence microscopy.
The multiple cloning site (MCS) in pEF1α-AcGFP1-N1 is positioned between the EF1 promoter (PEF1α) and the AcGFP1 coding sequence. Expression of the fusion protein is driven by the EF1α promoter, which remains constitutively active even after stable integration of the vector into the host cell genome (1). A Kozak consensus sequence, located immediately upstream of the AcGFP1 gene, enhances the translational efficiency of AcGFP1 in eukaryotic systems (2), and SV40 polyadenylation signals direct proper processing of the 3' end of the AcGFP1 mRNA.
The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418 (3). This cassette consists of the SV40 early promoter (PSV40 e), the Tn5 neomycin/kanamycin resistance gene, and polyadenylation signals from the herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of the cassette drives expression of the kanamycin resistance gene in E. coli.
Location of Features
PEF1α (human elongation factor 1 alpha promoter): 12–1346
MCS (multiple cloning site): 1348–1422
AcGFP1(human-codon-optimized): 1430–2146
SV40 polyA signal: 2302–2336
f1 origin of replication: 2399–2854 (complementary)
PSV40 e (SV40 early promoter and enhancer sequences): 3028–3296
SV40 origin of replication: 3195–3333
Kanr/Neor (kanamycin/neomycin resistance gene): 3379–4173
HSV TK polyA signals: 4409–4427
pUC origin of replication: 4758–5401
Additional Information
The gene of interest must be cloned into pEF1α-AcGFP1-N1 so that it is in-frame with the AcGFP1 coding sequence. Thegene must contain a start codon (ATG), and lack in-frame stop codons.
The pEF1α-AcGFP1-N1 vector can be transfected into mammalian cells using any standard transfection method. pEF1α-AcGFP1-N1 can be used as a cotransfection marker, as the unmodified vector will express AcGFP1 in mammalian cells.If required, stable transfectants can be selected using G418 (3).
Propagation in E. coli
Suitable host strains: DH5α, HB101 and other general purpose strains. Single-stranded DNA production requires
a host containing an F plasmid, such as the JM109 or XL1-Blue strains.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts.
E. coli replication origin: pUC
Copy number: high
Excitation and Emission Maxima of AcGFP1
Excitation: 475 nm
Emission: 505 nm
其他哺乳动物表达载体: