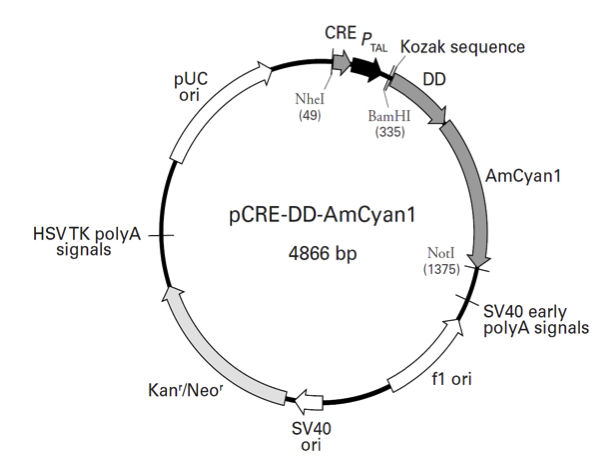
pCRE-DD-AmCyan1
pCRE-DD-AmCyan1
编号 | 载体名称 |
北京华越洋生物VECT6012 | pCRE-DD-AmCyan1 |
pCRE-DD-AmCyan1载体基本信息
载体名称: | pCRE-DD-AmCyan1 |
质粒类型: | 哺乳动物细胞表达载体;信号通路报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | TA(TATA like promoter) |
载体大小: | 4866 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | - |
载体标签: | 青色荧光蛋白DD-AmCyan1 |
载体抗性: | 卡那霉素 |
筛选标记: | 新霉素(Neomycin) |
克隆菌株: | DH5α, HB101 |
宿主细胞(系): | 常规细胞系,293、CV-1、CHO等 |
备注: | pCRE-DD-AmCyan1载体是cAMP信号通路报告载体,用来检测cAMP应答元件结合蛋白的活性; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
pCRE-DD-AmCyan1载体质粒图谱和多克隆位点信息
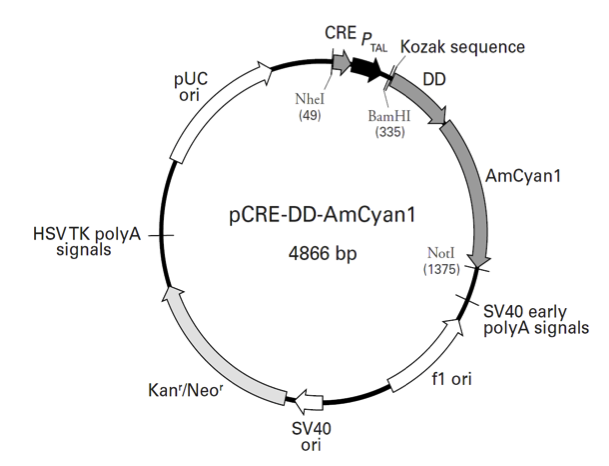
pCRE-DD-AmCyan1载体简介
pCRE-DD-AmCyan1 is a reporter vector that allows you to monitor cAMP response element binding protein (CREB) activation in mammalian cells. The vector contains two copies of the cAMP response element (CRE; 1) fused to a TATA-like promoter (PTAL) region from the herpes simplex virus thymidine kinase (HSV-TK) gene. The vector encodes the reporter protein DD-AmCyan1, a ligand-dependent, destabilized cyan fluorescent protein that minimizes background fluorescence from leaky promoters.
AmCyan1 is a human codon-optimized variant of the wild-type Anemonia majano cyan fluorescent protein (AmCyan) that exhibits enhanced emission characteristics (excitation and emission maxima: 458 and 489, respectively; 2, 3). DD-AmCyan1 is a modified version of AmCyan1 that is tagged on its N-terminus with the ProteoTuner destabilization domain (DD; 4). The presence of this destabilization domain causes rapid, proteasomal degradation of the fluorescent fusion protein; however, when the membrane permeant ligand Shield1 is added to the medium, it binds to the destabilization domain and protects the fusion protein from degradation.
In the absence of Shield1, the destabilization domain causes the degradation of any DD-AmCyan1 reporter protein produced prior to promoter activation, thus reducing background fluorescence. In order to analyze CREB activation, an inducer of choice is added to the medium along with the Shield1 stabilizing ligand, which effectively stabilizes the reporter protein, allowing it to accumulate. As a result, only the reporter molecules expressed during promoter induction will contribute to the fluorescence signal, providing a considerably higher signal-to-noise ratio than that obtained with non-destabilized or constitutively destabilized reporter systems. The high signal-to-noise ratio also allows the monitoring of CREB activation during discrete windows of time when Shield1 is added to the cell medium for discrete periods of time.
The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418 (5). This cassette consists of the SV40 early promoter, a Tn5 kanamycin/neomycin resistance gene, and herpes simplex virus thymidine kinase (HSV TK) polyadenylation signals. A bacterial promoter upstream of the cassette expresses kanamycin resistance in E. coli.
Use
The pCRE-DD-AmCyan1 Reporter vector, available as part of the CRE DD Cyan Reporter System , can be used to monitor CREB activation in live cells as well as in vivo. pCRE-DD-AmCyan1 can be transfected into mammalian cells using any standard transfection method. If required, stable transfectants can be selected using G418.
Propagation in E. coli Recommended host strains: DH5α, HB101, and other general purpose strains. Single-stranded DNA production requires a host containing an F plasmid such as JM109 or XL1-Blue. Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts. E. coli replication origin: pUC Copy number: high Plasmid incompatibility group: pMB1/ColE1
Excitation and emission maxima of AmCyan1 Excitation maximum = 458 nm Emission maximum = 489 nm
其他哺乳动物表达载体:
pcDNA6.2/nGeneBLAzer-GW/D-TOPO |