pcDNA6.2/nGeneBLAzer-GW/D-TOPO
pcDNA6.2/nGeneBLAzer-GW/D-TOPO
编号 | 载体名称 |
北京华越洋生物VECT6071 | pcDNA6.2/nGeneBLAzer-GW/D-TOPO |
pcDNA6.2/nGeneBLAzer-GW/D-TOPO载体基本信息
载体名称: | pcDNA6.2/nGeneBLAzer-GW/D-TOPO |
质粒类型: | 哺乳动物表达载体;β内酰胺酶报告载体;TOPO载体;cDNA表达载体; |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | TOPO-TA |
启动子: | CMV |
载体大小: | 5945 bp |
5' 测序引物及序列: | T7 Forward: 5’-TAATACGACTCACTATAGGG-3’ |
3' 测序引物及序列: | TK polyA Reverse: 5-TTGTCTCCTTCCGTGTTTCA-3 |
载体标签: | n-BLA (Beta-Lactamase) ; V5 (C Term) |
载体抗性: | 氨苄青霉素 |
筛选标记: | Blasticidin |
克隆菌株: | Mach1-T1R |
宿主细胞(系): | 常规细胞系,如293、Hela等 |
备注: | pcDNA6.2/nGeneBLAzer-GW/D-TOPO载体是β内酰胺酶报告载体; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | 组成型 |
病毒/非病毒: | 非病毒 |
pcDNA6.2/nGeneBLAzer-GW/D-TOPO载体质粒图谱和多克隆位点信息
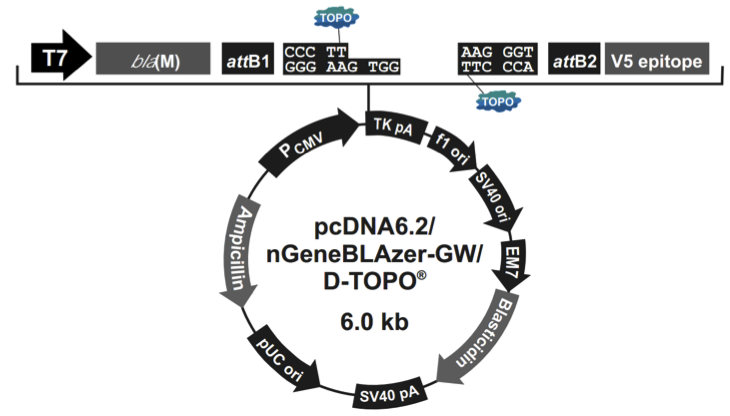

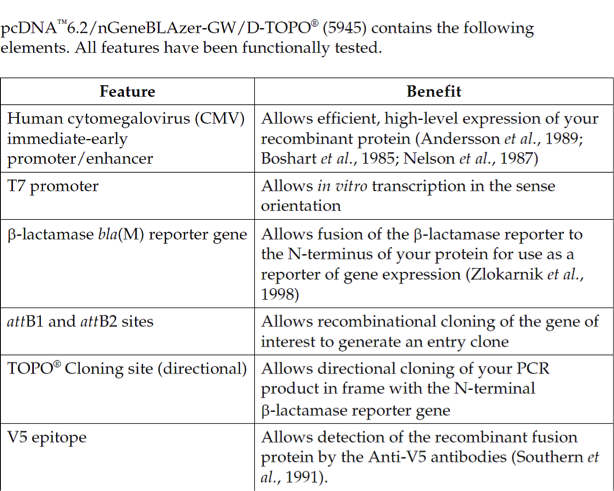
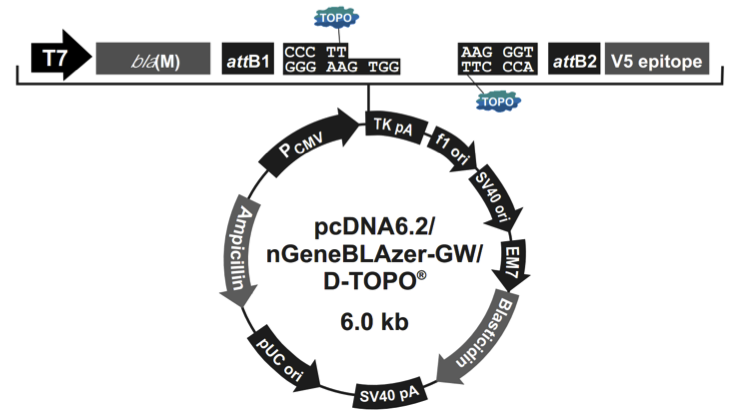
pcDNA6.2/nGeneBLAzer-GW/D-TOPO载体含有以下元件:
Human cytomegalovirus immediate-early (CMV) promoter/enhancer for high-level expression in a wide range of mammalian cells
β-lactamase bla(M) reporter gene for C-terminal (pcDNA6.2/cGeneBLAzerGW/D-TOPO) or N-terminal (pcDNA6.2/nGeneBLAzer-GW/D-TOPO)fusion to the gene of interest
attB1 and attB2 sites for site-specific recombination of the expression clone with a Gateway donor vector to generate an entry clone
Directional TOPO Cloning site for rapid and efficient directional cloning of blunt-end PCR products (see page 3 for more information)
The V5 epitope tag for detection using Anti-V5 antibodies (pcDNA6.2/nGeneBLAzer-GW/D-TOPO only)
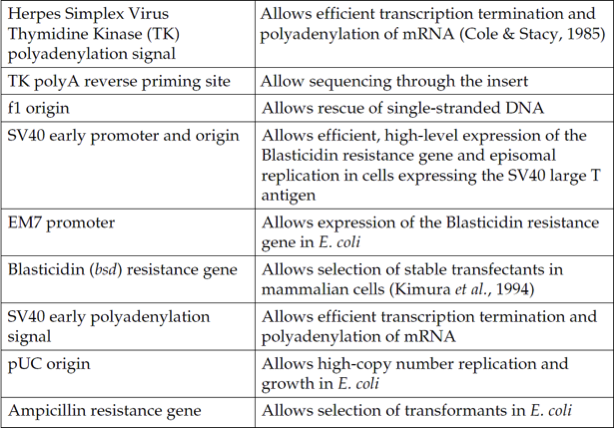
The Herpes Simplex Virus thymidine kinase polyadenylation signal for proper termination and processing of the recombinant transcript
f1 intergenic region for production of single-strand DNA in F plasmidcontaining E. coli
SV40 early promoter and origin for expression of the Blasticidin resistance gene and stable propagation of the plasmid in mammalian hosts expressing the SV40 large T antigen
Blasticidin resistance gene for selection of stable cell lines
The pUC origin for high copy replication and maintenance of the plasmid in E. coli
The ampicillin resistance gene for selection in E. coli
GeneBLAzer技术的优势
Suitable for use as a sensitive reporter of gene expression in living mammalian cells using fluorescence microscopy.
Provides ratiometric readout to minimize differences due to variability in cell number, substrate concentration, fluorescence intensity, and emission sensitivity.
Compatible with a wide variety of in vivo and in vitro applications including microplate-based transcriptional assays and flow cytometry.
Provides a flexible and simple assay development platform for gene expression in mammalian cells.
Using a non-toxic substrate allows continued cell culturing after quantitative analysis.
GeneBLAzer系统的组成
The GeneBLAzer System facilitates fluorescence detection of β-lactamase reporter activity in mammalian cells, and consists of two major components:
The β-lactamase reporter gene, bla(M), a truncated form of the E. coli bla gene.
When fused to a gene of interest, the bla(M) gene can be used as a reporter of gene expression in mammalian cells. For more information about the bla(M) gene, see below.
A fluorescence resonance energy transfer (FRET)-enabled substrate, CCF2 to facilitate fluorescence detection of β-lactamase activity. In the absence or presence of β-lactamase reporter activity, cells loaded with the CCF2 substrate fluoresce green or blue, respectively. Comparing the ratio of blue to green fluorescence in a population of live cells or in a cell extract of your sample to a negative control provides a means to quantitate gene expression. For more information about the CCF2 substrate and how FRET works, refer to the GeneBLAzer Detection Kits manual.
β内酰胺酶基因(bla)
β-lactamase is the product encoded by the ampicillin resistance gene (bla) and is the bacterial enzyme that hydrolyzes penicillins and cephalosporins. The bla gene is present in many cloning vectors and allows ampicillin selection in E. coli. β-lactamase enzyme activity is not found in mammalian cells.bla(M) Gene The GeneBLAzer Technology uses a modiied bla gene as a reporter in mammalian cells. This bla gene is derived from the E. coli TEM-1 gene present in many cloning vectors (Zlokarnik et al., 1998), and has been modified in the following ways:
72 nucleotides encoding the first 24 amino acids of β-lactamase were deleted from the N-terminal region of the gene. These 24 amino acids comprise the bacterial periplasmic signal sequence, and deleting this region allows cytoplasmic expression of β-lactamase in mammalian cells.
The amino acid at position 24 was mutated from His to Asp to create an optimal Kozak sequence for optimal translation initiation. This modified reporter gene is named bla(M).
Note: The TEM-1 gene also contains 2 mutations (at nucleotide positions 452 and 753) that distinguish it from the bla gene in pBR322 (Sutcliffe, 1978).
检测重组蛋白
Depending on the kit you are using, you will assay for β-lactamase reporter activity through in vivo or in vitro detection methods. A brief description of each detection method is provided below. For detailed information, refer to the GeneBLAzer Detection Kits manual. If you have generated a pcDNA6.2/nGeneBLAzer-GW/D-TOPO expression construct that contains your gene of interest fused to the V5 epitope tag, you may also detect your recombinant fusion protein by Western blot analysis using the Anti-V5 Antibodies.
体外检测
Using the GeneBLAzer In Vitro Detection Kit allows you to quantitate the amount of intracellular β-lactamase in cells based on the β-lactamase activity in lysates.
To detect β-lactamase activity in mammalian cell lysates, use the CCF2-FA substrate. CCF2-FA is the non-esterified, free acid form of CCF2, and is recommended for in vitro use because it is readily soluble in aqueous solution and may be added directly to pre-made cell lysates. Once added to cell lysates, you may quantitate the CCF2-FA fluorescence signal using a fluorescence plate reader or a fluorometer.
To prepare cell lysates from mammalian cells containing the bla(M) reporter gene, you must use a method that will preserve the activity of the β-lactamase enzyme.
Refer to the GeneBLAzer Detection Kits manual for detailed guidelines and protocols to prepare CCF2-FA solution, prepare cell lysates and samples, and detect CCF2 signal.
体内检测
Using the GeneBLAzer In Vivo Detection Kit allows you to measure β-lactamase reporter activity in live mammalian cells. Once β-lactamase reporter activity has been measured, cells may cultured further for use in additional assays or other downstream applications.
To detect β-lactamase activity in live mammalian cells, use the CCF2-AM substrate. CCF2-AM is the membrane-permeable, esterified form of CCF2, and is recommended for in vivo use because it is non-toxic, lipophilic, and readily enters the cell. Once cells are “loaded” with CCF2-AM, you may quantitate the CCF2 fluorescence signal using a variety of methods.
Refer to the GeneBLAzer Detection Kits manual for detailed guidelines and protocols to prepare CCF2-AM solution, load cells with CCF2-AM substrate, and detect CCF2 signal.
实验材料:
Purified plasmid DNA of your entry clone (50–150 ng/ μL in TE, pH 8.0)
pcDNA6.2/nGeneBLAzer-GW/D-TOPO or pcDNA6.2/nGeneBLAzer-GW/D-TOPO vector (150 ng/μL in TE, pH 8.0)
LR Clonase enzyme mix (keep at –80°C until immediately before use)
5X LR Clonase Reaction Buffer (supplied with the LR Clonase enzyme mix)
pENTR-gus positive control, optional (50 ng/μL in TE, pH 8.0; supplied with the LR Clonase enzyme mix)
TE Buffer, pH 8.0 (10 mM Tris-HCl, pH 8.0, 1 mM EDTA)
2 μg/μL Proteinase K solution (supplied with the LR Clonase enzyme mix; thaw and keep on ice until use)
Appropriate competent E. coli host and growth media for expression
S.O.C. Medium
LB agar plates containing the appropriate antibiotic to select for expression clones
pcDNA6.2/nGeneBLAzer-GW/D-TOPO载体序列
GACGGATCGGGAGATCTCCCGATCCCCTATGGTGCACTCTCAGTACAATCTGCTCTGATGCCGCATAGTT
AAGCCAGTATCTGCTCCCTGCTTGTGTGTTGGAGGTCGCTGAGTAGTGCGCGAGCAAAATTTAAGCTACA
ACAAGGCAAGGCTTGACCGACAATTGCATGAAGAATCTGCTTAGGGTTAGGCGTTTTGCGCTGCTTCGCG
ATGTACGGGCCAGATATACGCGTTGACATTGATTATTGACTAGTTATTAATAGTAATCAATTACGGGGTC
ATTAGTTCATAGCCCATATATGGAGTTCCGCGTTACATAACTTACGGTAAATGGCCCGCCTGGCTGACCG
CCCAACGACCCCCGCCCATTGACGTCAATAATGACGTATGTTCCCATAGTAACGCCAATAGGGACTTTCC
ATTGACGTCAATGGGTGGAGTATTTACGGTAAACTGCCCACTTGGCAGTACATCAAGTGTATCATATGCC
AAGTACGCCCCCTATTGACGTCAATGACGGTAAATGGCCCGCCTGGCATTATGCCCAGTACATGACCTTA
TGGGACTTTCCTACTTGGCAGTACATCTACGTATTAGTCATCGCTATTACCATGGTGATGCGGTTTTGGC
AGTACATCAATGGGCGTGGATAGCGGTTTGACTCACGGGGATTTCCAAGTCTCCACCCCATTGACGTCAA
TGGGAGTTTGTTTTGGCACCAAAATCAACGGGACTTTCCAAAATGTCGTAACAACTCCGCCCCATTGACG
CAAATGGGCGGTAGGCGTGTACGGTGGGAGGTCTATATAAGCAGAGCTCTCTGGCTAACTAGAGAACCCA
CTGCTTACTGGCTTATCGAAATTAATACGACTCACTATAGGGAGACCCAAGCTGGCTAGTTAAGCTGAGC
ATCAACAAGTTTGTACAAAAAAGCAGGCTCCGCGGCCGCCCCCTTCACCAAGGGTGGGCGCGCCGACCCA
GCTTTCTTGTACAAAGTGGTTGATGCTGTTATGGACCCAGAAACGCTGGTGAAAGTAAAAGATGCTGAAG
ATCAGTTGGGTGCACGAGTGGGTTACATCGAACTGGATCTCAACAGCGGTAAGATCCTTGAGAGTTTTCG
CCCCGAAGAACGTTTTCCAATGATGAGCACTTTTAAAGTTCTGCTATGTGGCGCGGTATTATCCCGTATT
GACGCCGGGCAAGAGCAACTCGGTCGCCGCATACACTATTCTCAGAATGACTTGGTTGAGTACTCACCAG
TCACAGAAAAGCATCTTACGGATGGCATGACAGTAAGAGAATTATGCAGTGCTGCCATAACCATGAGTGA
TAACACTGCGGCCAACTTACTTCTGACAACGATCGGAGGACCGAAGGAGCTAACCGCTTTTTTGCACAAC
ATGGGGGATCATGTAACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATACCAAACGACGAGC
GTGACACCACGATGCCTGTAGCAATGGCAACAACGTTGCGCAAACTATTAACTGGCGAACTACTTACTCT
AGCTTCCCGGCAACAATTAATAGACTGGATGGAGGCGGATAAAGTTGCAGGACCACTTCTGCGCTCGGCC
CTTCCGGCTGGCTGGTTTATTGCTGATAAATCTGGAGCCGGTGAGCGTGGGTCTCGCGGTATCATTGCAG
CACTGGGGCCAGATGGTAAGCCCTCCCGTATCGTAGTTATCTACACGACGGGGAGTCAGGCAACTATGGA
TGAACGAAATAGACAGATCGCTGAGATAGGTGCCTCACTGATTAAGCATTGGTAACCGGTTAGTAATGAG
TTTAAACGGGGGAGGCTAACTGAAACACGGAAGGAGACAATACCGGAAGGAACCCGCGCTATGACGGCAA
TAAAAAGACAGAATAAAACGCACGGGTGTTGGGTCGTTTGTTCATAAACGCGGGGTTCGGTCCCAGGGCT
GGCACTCTGTCGATACCCCACCGAGACCCCATTGGGGCCAATACGCCCGCGTTTCTTCCTTTTCCCCACC
CCACCCCCCAAGTTCGGGTGAAGGCCCAGGGCTCGCAGCCAACGTCGGGGCGGCAGGCCCTGCCATAGCA
GATCTGCGCAGCTGGGGCTCTAGGGGGTATCCCCACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTG
TGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCC
TTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGCATCCCTTTAGGGTTCCGA
TTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTGATGGTTCACGTAGTGGGCCATCGC
CCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAAC
TGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGATTTTGGGGATTTCGGCCTAT
TGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTAATTCTGTGGAATGTGTGTCAGTTAGG
GTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACC
AGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAA
CCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCA
TGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTCCAGAAGTAG
TGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAAAGCTCCCGGGAGCTTGTATATCCATTTTCGGAT
CTGATCAGCACGTGTTGACAATTAATCATCGGCATAGTATATCGGCATAGTATAATACGACAAGGTGAGG
AACTAAACCATGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAGAGCAACGGCTACAATCA
ACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGCGACGGCCGCATCTTCAC
TGGTGTCAATGTATATCATTTTACTGGGGGACCTTGTGCAGAACTCGTGGTGCTGGGCACTGCTGCTGCT
GCGGCAGCTGGCAACCTGACTTGTATCGTCGCGATCGGAAATGAGAACAGGGGCATCTTGAGCCCCTGCG
GACGGTGCCGACAGGTGCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTGAAGGACAGTGATGGACA
GCCGACGGCAGTTGGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGGGCTAAGCACTTCGTGGC
CGAGGAGCAGGACTGACACGTGCTACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTC
GGAATCGTTTTCCGGGACGCCGGCTGGATGATCCTCCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCC
ACCCCAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAA
AGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTATCATGTCTGTATA
CCGTCGACCTCTAGCTAGAGCTTGGCGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATTGTTATCCGC
TCACAATTCCACACAACATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTA
ACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAA
TGAATCGGCCAACGCGCGGGGAGAGGCGGTTTGCGTATTGGGCGCTCTTCCGCTTCCTCGCTCACTGACT
CGCTGCGCTCGGTCGTTCGGCTGCGGCGAGCGGTATCAGCTCACTCAAAGGCGGTAATACGGTTATCCAC
AGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAG
GCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTC
AGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTC
TCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCT
CATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAAC
CCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGA
CTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAG
TTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGC
CAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTTTTTT
TGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGG
TCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCA
CCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGA
CAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCC
TGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATAC
CGCGAGACCCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAG
AAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGT
TCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTG
GTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAA
AGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTT
ATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACT
CAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAA
TACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCA
AGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTT
TTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGC
GACACGGAAATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGT
CTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCC
GAAAAGTGCCACCTGACGTC
其他哺乳动物表达载体: