pZsGreen1-DR
pZsGreen1-DR
编号 | 载体名称 |
北京华越洋VECT75377 | pZsGreen1-DR |
pZsGreen1-DR载体基本信息:
载体名称: | pZsGreen1-DR |
质粒类型: | 无启动子载体;荧光报告载体 |
高拷贝/低拷贝: | 高拷贝 |
克隆方法: | 限制性内切酶,多克隆位点 |
启动子: | 无启动子 |
载体大小: | 4251 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | 绿色荧光蛋白ZsGreen1-DR |
载体抗性: | 卡那霉素 |
筛选标记: | Neomycin_1.html' target='_blank'>新霉素(Neomycin) |
克隆菌株: | DH5α, JM109等 |
宿主细胞(系): | 原核细胞和真核细胞均可 |
备注: | pZsGreen1-DR载体不含启动子,将待研究的目的启动子插入MCS区,用于研究启动子及顺式转录元件在细胞内的转录水平和活性; |
稳定性: | 瞬表达 或 稳表达 |
组成型/诱导型: | -- |
病毒/非病毒: | 非病毒 |
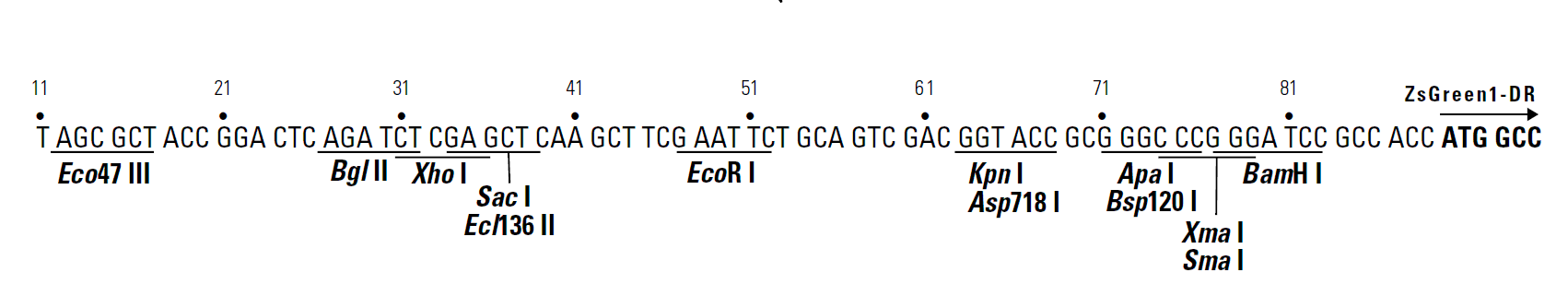
pZsGreen1-DR载体质粒图谱和多克隆位点信息


pZsGreen1-DR载体描述:
pZsGreen1-DR is a promoterless vector that encodes ZsGreen1-DR, a destabilized variant of
Zoanthus sp. green fluorescent protein (ZsGreen; 1). The ZsGreen1 coding sequence contains
a series of silent base-pair changes, which correspond to human codon-usage preferences, for
optimal expression in mammalian cells (2). In contrast to the original protein, ZsGreen1-DR has a
short half-life, making it well suited for studies that require rapid reporter turnover. This destabilized
variant was constructed by fusing the C-terminus of the protein to amino acid residues 422–461 of
mouse ornithine decarboxylase (MODC), one of the most short-lived proteins in mammalian cells (3).
This region of MODC contains a PEST sequence that targets the protein for degradation, resulting
in rapid protein turnover (3,4). Three point mutations in this sequence further reduce the protein
half-life (3). Sequences upstream of ZsGreen1-DR have been converted to a Kozak consensus
translation initiation site (5) to enhance translation efficiency in eukaryotic cells. A single amino acid
substitution (Asn-65 to Met) has been made to enhance the emission characteristics of ZsGreen1
(excitation maximum = 496 nm; emission maximum = 506 nm).
pZsGreen1-DR can be used to monitor transcription from different promoters and promoter/enhancer
combinations inserted into the multiple cloning site (MCS), located upstream of the ZsGreen1-DR
coding sequence. Downstream SV40 polyadenylation signals direct proper processing of the 3'
end of the ZsGreen1-DR mRNA. The vector backbone contains an SV40 origin for replication in
mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in
E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor)
allows stably transfected eukaryotic cells to be selected using G418. This cassette consists of the
SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals
from the Herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of
the cassette expresses kanamycin resistance in E. coli.
pZsGreen1-DR载体使用:
ZsGreen1-DR can be used as an in vivo reporter of gene expression. Because of its rapid turnover rate, its expression from a promoter
of interest provides a more accurate assessment of the promoter's activity over time than does the more stable ZsGreen1.
Promoter/enhancer elements should be inserted in the MCS upstream of the ZsGreen1-DR coding sequence. Without the addition of a
functional promoter, this vector will not express ZsGreen1-DR. The recombinant pZsGreen1-DR vector can be transfected into mammalian
cells using any standard transfection method. If required, stable transformants can be selected using G418 (6).
Location of features
MCS: 12–83
Destabilized Zoanthus sp. Green Fluorescent Protein (ZsGreen1-DR) gene
Kozak consensus translation initiation site: 83–93
Start codon (ATG): 90–92; Stop codon: 915–917
Asn-65 to Met mutation (A→T, C→G ): 286, 287
Mouse ornithine decarboxylase PEST sequence: 795–917
SV40 early mRNA polyadenylation signal
Polyadenylation signals: 1070–1075 & 1099–1104
mRNA 3' ends: 1108 & 1120
f1 single-strand DNA origin: 1167–1622
(Packages noncoding strand of ZsGreen1-DR.)
Ampicillin resistance (β-lactamase) promoter
–35 region: 1684–1689; –10 region: 1707–1712
Transcription start point: 1719
SV40 origin of replication: 1963–2098
SV40 early promoter
Enhancer (72-bp tandem repeats): 1794–1867 & 1868–1939
21-bp repeats: 1943–1963, 1964–1984 & 1986–2006
Early promoter element: 2019–2025
Major transcription start points: 2015, 2053, 2059 & 2064
Kanamycin/neomycin resistance geneNeomycin phosphotransferase coding sequences:
Start codon (ATG): 2147–2149; stop codon: 2939–2941
G→A mutation to remove Pst I site: 2329
C→A (Arg→Ser) mutation to remove BssH II site: 2675
Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal
Polyadenylation signals: 3177–3182 & 3190–3195
pUC plasmid replication origin: 3526–4169
Excitation and emission maxima of ZsGreen1
Excitation maximum = 496 nm
Emission maximum = 506 nm
pZsGreen1-DR载体序列:
ORIGIN
1 TAGTTATTAC TAGCGCTACC GGACTCAGAT CTCGAGCTCA AGCTTCGAAT TCTGCAGTCG
61 ACGGTACCGC GGGCCCGGGA TCCGCCACCA TGGCCCAGTC CAAGCACGGC CTGACCAAGG
121 AGATGACCAT GAAGTACCGC ATGGAGGGCT GCGTGGACGG CCACAAGTTC GTGATCACCG
181 GCGAGGGCAT CGGCTACCCC TTCAAGGGCA AGCAGGCCAT CAACCTGTGC GTGGTGGAGG
241 GCGGCCCCTT GCCCTTCGCC GAGGACATCT TGTCCGCCGC CTTCATGTAC GGCAACCGCG
301 TGTTCACCGA GTACCCCCAG GACATCGTCG ACTACTTCAA GAACTCCTGC CCCGCCGGCT
361 ACACCTGGGA CCGCTCCTTC CTGTTCGAGG ACGGCGCCGT GTGCATCTGC AACGCCGACA
421 TCACCGTGAG CGTGGAGGAG AACTGCATGT ACCACGAGTC CAAGTTCTAC GGCGTGAACT
481 TCCCCGCCGA CGGCCCCGTG ATGAAGAAGA TGACCGACAA CTGGGAGCCC TCCTGCGAGA
541 AGATCATCCC CGTGCCCAAG CAGGGCATCT TGAAGGGCGA CGTGAGCATG TACCTGCTGC
601 TGAAGGACGG TGGCCGCTTG CGCTGCCAGT TCGACACCGT GTACAAGGCC AAGTCCGTGC
661 CCCGCAAGAT GCCCGACTGG CACTTCATCC AGCACAAGCT GACCCGCGAG GACCGCAGCG
721 ACGCCAAGAA CCAGAAGTGG CACCTGACCG AGCACGCCAT CGCCTCCGGC TCCGCCTTGC
781 CCAAGCTTCC GCGGAGCCAT GGCTTCCCGC CGGCGGTGGC GGCGCAGGAT GATGGCACGC
841 TGCCCATGTC TTGTGCCCAG GAGAGCGGGA TGGACCGTCA CCCTGCAGCC TGTGCTTCTG
901 CTAGGATCAA TGTGTAGGCG GCCGCGACTC TAGATCATAA TCAGCCATAC CACATTTGTA
961 GAGGTTTTAC TTGCTTTAAA AAACCTCCCA CACCTCCCCC TGAACCTGAA ACATAAAATG
1021 AATGCAATTG TTGTTGTTAA CTTGTTTATT GCAGCTTATA ATGGTTACAA ATAAAGCAAT
1081 AGCATCACAA ATTTCACAAA TAAAGCATTT TTTTCACTGC ATTCTAGTTG TGGTTTGTCC
1141 AAACTCATCA ATGTATCTTA AGGCGTAAAT TGTAAGCGTT AATATTTTGT TAAAATTCGC
1201 GTTAAATTTT TGTTAAATCA GCTCATTTTT TAACCAATAG GCCGAAATCG GCAAAATCCC
1261 TTATAAATCA AAAGAATAGA CCGAGATAGG GTTGAGTGTT GTTCCAGTTT GGAACAAGAG
1321 TCCACTATTA AAGAACGTGG ACTCCAACGT CAAAGGGCGA AAAACCGTCT ATCAGGGCGA
1381 TGGCCCACTA CGTGAACCAT CACCCTAATC AAGTTTTTTG GGGTCGAGGT GCCGTAAAGC
1441 ACTAAATCGG AACCCTAAAG GGAGCCCCCG ATTTAGAGCT TGACGGGGAA AGCCGGCGAA
1501 CGTGGCGAGA AAGGAAGGGA AGAAAGCGAA AGGAGCGGGC GCTAGGGCGC TGGCAAGTGT
1561 AGCGGTCACG CTGCGCGTAA CCACCACACC CGCCGCGCTT AATGCGCCGC TACAGGGCGC
1621 GTCAGGTGGC ACTTTTCGGG GAAATGTGCG CGGAACCCCT ATTTGTTTAT TTTTCTAAAT
1681 ACATTCAAAT ATGTATCCGC TCATGAGACA ATAACCCTGA TAAATGCTTC AATAATATTG
1741 AAAAAGGAAG AGTCCTGAGG CGGAAAGAAC CAGCTGTGGA ATGTGTGTCA GTTAGGGTGT
1801 GGAAAGTCCC CAGGCTCCCC AGCAGGCAGA AGTATGCAAA GCATGCATCT CAATTAGTCA
1861 GCAACCAGGT GTGGAAAGTC CCCAGGCTCC CCAGCAGGCA GAAGTATGCA AAGCATGCAT
1921 CTCAATTAGT CAGCAACCAT AGTCCCGCCC CTAACTCCGC CCATCCCGCC CCTAACTCCG
1981 CCCAGTTCCG CCCATTCTCC GCCCCATGGC TGACTAATTT TTTTTATTTA TGCAGAGGCC
2041 GAGGCCGCCT CGGCCTCTGA GCTATTCCAG AAGTAGTGAG GAGGCTTTTT TGGAGGCCTA
2101 GGCTTTTGCA AAGATCGATC AAGAGACAGG ATGAGGATCG TTTCGCATGA TTGAACAAGA
2161 TGGATTGCAC GCAGGTTCTC CGGCCGCTTG GGTGGAGAGG CTATTCGGCT ATGACTGGGC
2221 ACAACAGACA ATCGGCTGCT CTGATGCCGC CGTGTTCCGG CTGTCAGCGC AGGGGCGCCC
2281 GGTTCTTTTT GTCAAGACCG ACCTGTCCGG TGCCCTGAAT GAACTGCAAG ACGAGGCAGC
2341 GCGGCTATCG TGGCTGGCCA CGACGGGCGT TCCTTGCGCA GCTGTGCTCG ACGTTGTCAC
2401 TGAAGCGGGA AGGGACTGGC TGCTATTGGG CGAAGTGCCG GGGCAGGATC TCCTGTCATC
2461 TCACCTTGCT CCTGCCGAGA AAGTATCCAT CATGGCTGAT GCAATGCGGC GGCTGCATAC
2521 GCTTGATCCG GCTACCTGCC CATTCGACCA CCAAGCGAAA CATCGCATCG AGCGAGCACG
2581 TACTCGGATG GAAGCCGGTC TTGTCGATCA GGATGATCTG GACGAAGAGC ATCAGGGGCT
2641 CGCGCCAGCC GAACTGTTCG CCAGGCTCAA GGCGAGCATG CCCGACGGCG AGGATCTCGT
2701 CGTGACCCAT GGCGATGCCT GCTTGCCGAA TATCATGGTG GAAAATGGCC GCTTTTCTGG
2761 ATTCATCGAC TGTGGCCGGC TGGGTGTGGC GGACCGCTAT CAGGACATAG CGTTGGCTAC
2821 CCGTGATATT GCTGAAGAGC TTGGCGGCGA ATGGGCTGAC CGCTTCCTCG TGCTTTACGG
2881 TATCGCCGCT CCCGATTCGC AGCGCATCGC CTTCTATCGC CTTCTTGACG AGTTCTTCTG
2941 AGCGGGACTC TGGGGTTCGA AATGACCGAC CAAGCGACGC CCAACCTGCC ATCACGAGAT
3001 TTCGATTCCA CCGCCGCCTT CTATGAAAGG TTGGGCTTCG GAATCGTTTT CCGGGACGCC
3061 GGCTGGATGA TCCTCCAGCG CGGGGATCTC ATGCTGGAGT TCTTCGCCCA CCCTAGGGGG
3121 AGGCTAACTG AAACACGGAA GGAGACAATA CCGGAAGGAA CCCGCGCTAT GACGGCAATA
3181 AAAAGACAGA ATAAAACGCA CGGTGTTGGG TCGTTTGTTC ATAAACGCGG GGTTCGGTCC
3241 CAGGGCTGGC ACTCTGTCGA TACCCCACCG AGACCCCATT GGGGCCAATA CGCCCGCGTT
3301 TCTTCCTTTT CCCCACCCCA CCCCCCAAGT TCGGGTGAAG GCCCAGGGCT CGCAGCCAAC
3361 GTCGGGGCGG CAGGCCCTGC CATAGCCTCA GGTTACTCAT ATATACTTTA GATTGATTTA
3421 AAACTTCATT TTTAATTTAA AAGGATCTAG GTGAAGATCC TTTTTGATAA TCTCATGACC
3481 AAAATCCCTT AACGTGAGTT TTCGTTCCAC TGAGCGTCAG ACCCCGTAGA AAAGATCAAA
3541 GGATCTTCTT GAGATCCTTT TTTTCTGCGC GTAATCTGCT GCTTGCAAAC AAAAAAACCA
3601 CCGCTACCAG CGGTGGTTTG TTTGCCGGAT CAAGAGCTAC CAACTCTTTT TCCGAAGGTA
3661 ACTGGCTTCA GCAGAGCGCA GATACCAAAT ACTGTCCTTC TAGTGTAGCC GTAGTTAGGC
3721 CACCACTTCA AGAACTCTGT AGCACCGCCT ACATACCTCG CTCTGCTAAT CCTGTTACCA
3781 GTGGCTGCTG CCAGTGGCGA TAAGTCGTGT CTTACCGGGT TGGACTCAAG ACGATAGTTA
3841 CCGGATAAGG CGCAGCGGTC GGGCTGAACG GGGGGTTCGT GCACACAGCC CAGCTTGGAG
3901 CGAACGACCT ACACCGAACT GAGATACCTA CAGCGTGAGC TATGAGAAAG CGCCACGCTT
3961 CCCGAAGGGA GAAAGGCGGA CAGGTATCCG GTAAGCGGCA GGGTCGGAAC AGGAGAGCGC
4021 ACGAGGGAGC TTCCAGGGGG AAACGCCTGG TATCTTTATA GTCCTGTCGG GTTTCGCCAC
4081 CTCTGACTTG AGCGTCGATT TTTGTGATGC TCGTCAGGGG GGCGGAGCCT ATGGAAAAAC
4141 GCCAGCAACG CGGCCTTTTT ACGGTTCCTG GCCTTTTGCT GGCCTTTTGC TCACATGTTC
4201 TTTCCTGCGT TATCCCCTGA TTCTGTGGAT AACCGTATTA CCGCCATGCA T
//
pZsGreen1-DR载体其他相关的其他类型载体:

