pHIS1525
pHIS1525
编号 | 载体名称 |
北京华越洋VECT75310 | pHIS1525 |
pHIS1525载体基本信息:
载体名称: | pHIS1525 |
质粒类型: | 巨大芽孢杆菌表达载体 |
高拷贝/低拷贝: | -- |
启动子: | -- |
克隆方法: | 多克隆位点,限制性内切酶 |
载体大小: | 7402 bp |
5' 测序引物及序列: | -- |
3' 测序引物及序列: | -- |
载体标签: | 6xHis |
载体抗性: | 氨苄青霉素和四环素 |
筛选标记: | 四环素 |
备注: | -- |
稳定性: | -- |
组成型: | -- |
病毒/非病毒: | -- |
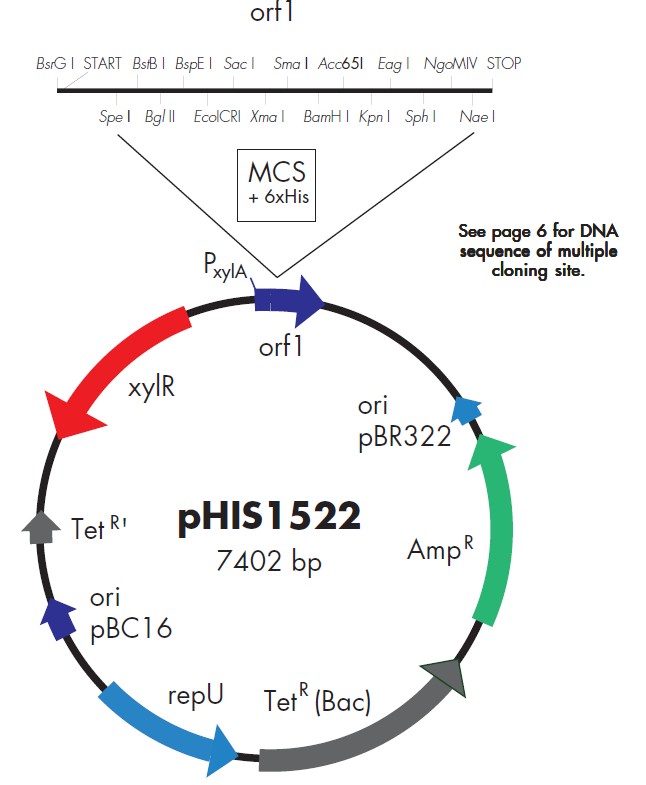
pHIS1525载体质粒图谱和多克隆位点信息:
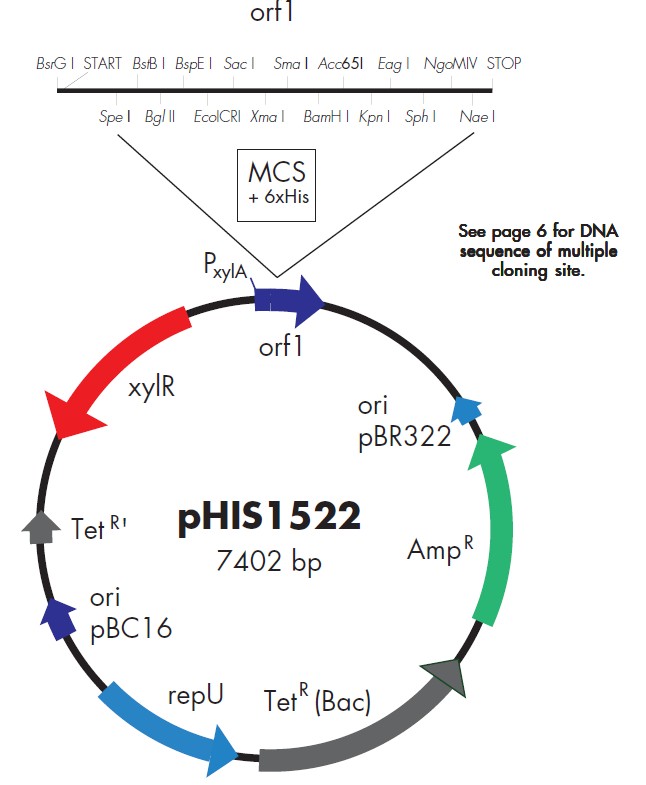
pHIS1525载体简介:
Plasmids pHIS1522/pHIS1525 both contain the strong xylA promoter (PxylA)
originating from Bacillus megaterium. Transcription from this promoter is xylose
inducible (after xylose addition, the xylose repressor coded by the xylR gene on
the plasmids is released from PxylA and transcription is initiated). The most
convenient cloning sites for insertion of DNA fragments carrying heterologous
genes are located in an open reading frame (orf1) under control of the xyloseinducible
promoter PxylA (15 unique restriction sites; see sequence). Therefore, any
protein can be expressed using one out of three functionally different fusion
strategies.
A transcriptional fusion requires that the gene of interest carries its own ribosome
binding sequence (RBS) and translation initiation codon. Such a DNA fragment
can be fused into any of the available restriction sites within orf1. Whether the
resulting transcriptional fusion leads to expression of the gene of interest, which
is independent from orf1 expression, depends on the location of the created orf1
stop codon with respect to the start codon of the gene of interest. lf these are close
together, translational coupling may occur, in which the ribosomes translating the
orf1 reading frame would terminate at its stop codon, creating a locally high
concentration of ribosomes, so that translation initiation at the new start codon
would be more efficient compared to a construct in which the orf1 translation
terminates farther away from the start codon.
On the other hand, if the orf1 reading frame continues for a long distance into the
reading frame of the gene of interest, the ribosomes translating the created orf1
fusion protein might inhibit initiation of translation of the protein of interest.
Therefore, it is advisable to pay attention to placement of a stop codon when
constructing the gene fusion. Taken together, although a transcriptional or operon
fusion is constructed, the efficient translation of the orf1 reading frame, and any
fusion thereof created by the insertion, is likely to, positively or negatively,
influence the translation efficiency of the gene of interest.
Alternatively, a truncated version of the gene of interest, lacking its own start
codon, may be fused in frame to the orf1 reading frame on pHIS1522/pHIS1525
to create a translational or protein fusion. This will result in expression of a chimeric
protein consisting of up to 18 amino acids specified by the orf1 encoding
sequence, followed by the sequence encoded by the gene of interest.
Using the BsrGI restriction site directly before the ATG start codon enables cloning
without changing the N-terminus of the protein of interest. pHIS1525 contains a
sequence encoding the B. subtilis extracellular esterase (LipA) and allows cloning
of target genes directly after the signal peptidase restriction site using the Kas I,
Nar I, or SfoI restriction sites.
It is important to note that the multiple cloning site and its reading frame are
identical in pHIS1522 and pHIS1525 starting from BglII. Hence, a parallel
cloning strategy of the gene of interest in pHIS1522 for intracellular and in
pHIS1525 for extracellular production is possible. The 6xHis sequence upstream
the MCS allows convenient purification and detection of the expressed Histagged
target proteins. For expression of target proteins without a 6xHis tag,
suitable plasmid constructs (pMM1522 & pMM1525) are also available.
pHIS1525载体其他相关的其他类型载体: