pDsRed2-1
pDsRed2-1
编号 | 载体名称 |
北京华越洋VECT75327 | pDsRed2-1 |
pDsRed2-1载体基本信息:
载体名称: | pDsRed2-1 |
质粒类型: | 无启动子载体;荧光报告载体 |
高拷贝/低拷贝: | -- |
启动子: | 无 |
克隆方法: | 多克隆位点,限制性内切酶 |
载体大小: | 4107bp |
5' 测序引物及序列: | 5'-AGCTGGACATCACCTCCCACAACG-3' |
3' 测序引物及序列: | -- |
载体标签: | 红色荧光蛋白DsRed2 |
载体抗性: | 卡那霉素(Kanamycin) |
筛选标记: | Neomycin_1.html' target='_blank'>新霉素(Neomycin) |
克隆菌株: | TOP10, DH5α,HB101 |
宿主细胞(系): | 真核细胞 & 原核细胞 |
备注: | pDsRed2-1载体不含启动子,将待研究的目的启动子插入MCS区,用于研究启动子及顺式转录元件在细胞内的作用水平 。 |
稳定性: | 稳表达 或 瞬表达 |
组成型: | -- |
病毒/非病毒: | 非病毒 |
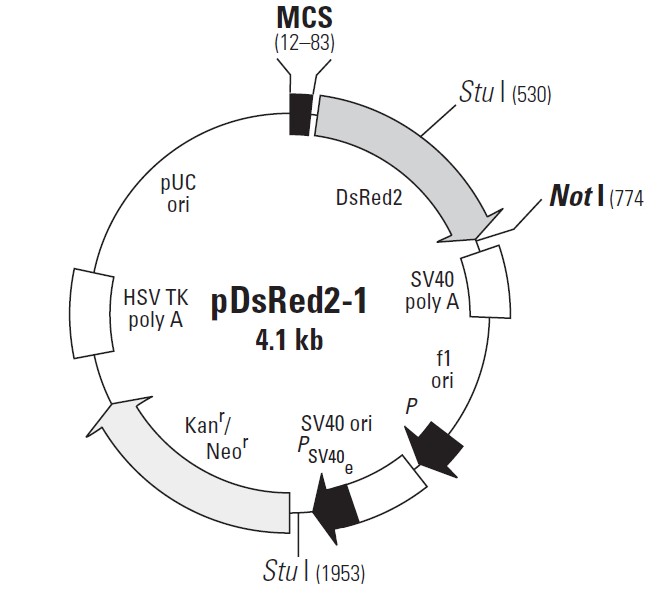
pDsRed2-1载体质粒图谱和多克隆位点信息:
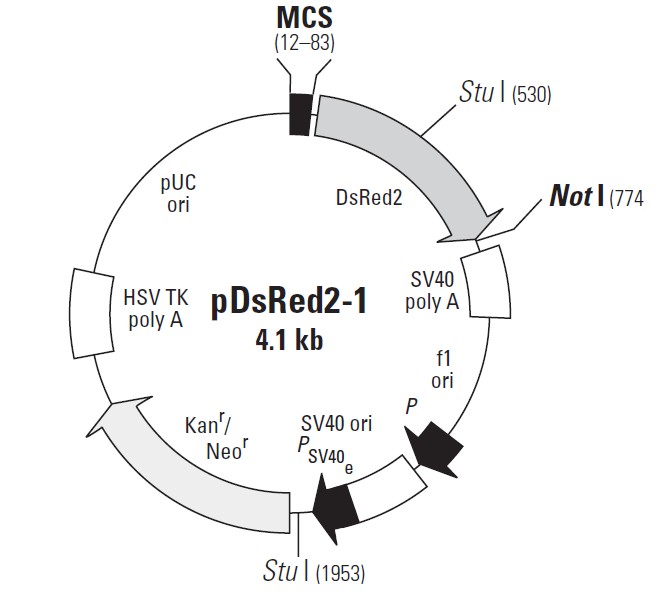
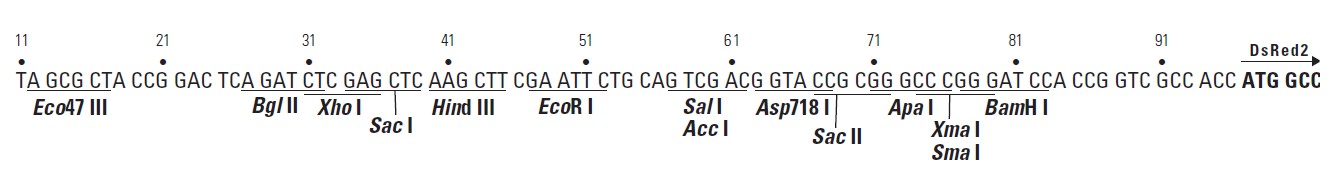
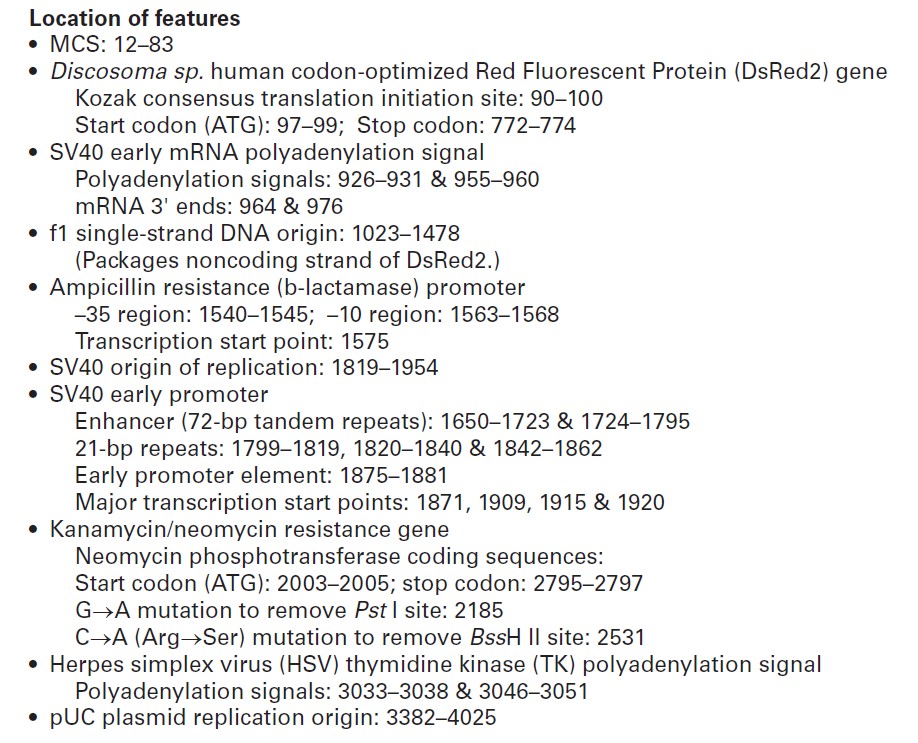
pDsRed2-1载体描述:
pDsRed2-1 encodes DsRed2, a DsRed variant engineered for faster maturation and lower non-specific aggregation. Derived from the Discosoma sp. red fluorescent protein (drFP583; 1), DsRed2, like its progenitor DsRed1, contains a series of silent base-pair changes that correspond to human codon-usage preferences for high expression in mammalian cells (2). In addition to these changes, DsRed2 contains six amino acid substitutions: V105A, I161T, and S197A, which result in the more rapid appearance of red fluorescence in transfected cell lines; and R2A, K5E, and K9T, which prevent the protein from aggregating. (DsRed2 may, however, form the same tetrameric structure as DsRed1 [3].) In mammalian cell cultures when DsRed2 is expressed constitutively, red-emitting cells can be detected by fluorescence microscopy within 24 hours of transfection. Large insoluble aggregates of protein, often observed in bacterial and mammalian cell systems expressing DsRed1, are dramatically reduced in cells expressing DsRed2. The faster-maturing, more soluble red fluorescent protein is also well tolerated by host cells; mammalian cell cultures transfected with DsRed2 show no obvious signs of reduced viability—in those cell lines tested, cells expressing DsRed2 display the same morphology (e.g., adherence, light-refraction) and growth characteristics as non-transfected controls.
pDsRed2-1 is a promoterless DsRed2 vector that can be used to monitor transcription from different promoters and promoter/enhancer combinations inserted into the multiple cloning site (MCS). Sequences upstream of DsRed2 have been converted to a Kozak consensus translation initiation site (4) to increase translation efficiency in eukaryotic cells. SV40 polyadenylation signals downstream of the DsRed2 gene direct proper processing of the 3' end of the DsRed2 mRNA. The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418. This cassette consists of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of the cassette expresses kanamycin resistance in E. coli.
DsRed2 can be used as an in vivo reporter of gene expression. Promoters should be cloned into the pDsRed2-1 MCS upstream from the DsRed2 coding sequence. Without addition of a functional promoter, this vector will not express DsRed2. The recombinant DsRed2 vector can be transfected into mammalian cells using any standard transfection method. If required, stable transfectants can be selected using G418 (5).
pDsRed2-1载体含有以下元件:
MCS: 12–83
Discosoma sp. human codon-optimized Red Fluorescent Protein (DsRed2) gene
Kozak consensus translation initiation site: 90–100
Start codon (ATG): 97–99; Stop codon: 772–774
SV40 early mRNA polyadenylation signal
Polyadenylation signals: 926–931 & 955–960
mRNA 3' ends: 964 & 976
f1 single-strand DNA origin: 1023–1478
(Packages noncoding strand of DsRed2.)
Ampicillin resistance (b-lactamase) promoter
–35 region: 1540–1545; –10 region: 1563–1568
Transcription start point: 1575
SV40 origin of replication: 1819–1954
SV40 early promoter
Enhancer (72-bp tandem repeats): 1650–1723 & 1724–1795
21-bp repeats: 1799–1819, 1820–1840 & 1842–1862
Early promoter element: 1875–1881
Major transcription start points: 1871, 1909, 1915 & 1920
Kanamycin/neomycin resistance geneNeomycin phosphotransferase coding sequences:
Start codon (ATG): 2003–2005; stop codon: 2795–2797
G→A mutation to remove Pst I site: 2185
C→A (Arg→Ser) mutation to remove BssH II site: 2531
Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signal
Polyadenylation signals: 3033–3038 & 3046–3051
pUC plasmid replication origin: 3382–4025
Propagation in E. Coli
Suitable host strains: DH5α, HB101 and other general purpose strains. Single-stranded DNA production requires
a host containing an F plasmid such as JM109 or XL1-Blue.
Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) to E. coli hosts.
E. coli replication origin: pUC
Copy number: ≈500
Plasmid incompatibility group: pMB1/Col E1
Red Fluorescent Protein (DsRed2)
Excitation/Emission Maxima: 558 nm / 583 nm
pDsRed2-1载体序列:
ORIGIN
1 TAGTTATTAC TAGCGCTACC GGACTCAGAT CTCGAGCTCA AGCTTCGAAT TCTGCAGTCG
61 ACGGTACCGC GGGCCCGGGA TCCACCGGTC GCCACCATGG CCTCCTCCGA GAACGTCATC
121 ACCGAGTTCA TGCGCTTCAA GGTGCGCATG GAGGGCACCG TGAACGGCCA CGAGTTCGAG
181 ATCGAGGGCG AGGGCGAGGG CCGCCCCTAC GAGGGCCACA ACACCGTGAA GCTGAAGGTG
241 ACCAAGGGCG GCCCCCTGCC CTTCGCCTGG GACATCCTGT CCCCCCAGTT CCAGTACGGC
301 TCCAAGGTGT ACGTGAAGCA CCCCGCCGAC ATCCCCGACT ACAAGAAGCT GTCCTTCCCC
361 GAGGGCTTCA AGTGGGAGCG CGTGATGAAC TTCGAGGACG GCGGCGTGGC GACCGTGACC
421 CAGGACTCCT CCCTGCAGGA CGGCTGCTTC ATCTACAAGG TGAAGTTCAT CGGCGTGAAC
481 TTCCCCTCCG ACGGCCCCGT GATGCAGAAG AAGACCATGG GCTGGGAGGC CTCCACCGAG
541 CGCCTGTACC CCCGCGACGG CGTGCTGAAG GGCGAGACCC ACAAGGCCCT GAAGCTGAAG
601 GACGGCGGCC ACTACCTGGT GGAGTTCAAG TCCATCTACA TGGCCAAGAA GCCCGTGCAG
661 CTGCCCGGCT ACTACTACGT GGACGCCAAG CTGGACATCA CCTCCCACAA CGAGGACTAC
721 ACCATCGTGG AGCAGTACGA GCGCACCGAG GGCCGCCACC ACCTGTTCCT GTAGCGGCCG
781 CGACTCTAGA TCATAATCAG CCATACCACA TTTGTAGAGG TTTTACTTGC TTTAAAAAAC
841 CTCCCACACC TCCCCCTGAA CCTGAAACAT AAAATGAATG CAATTGTTGT TGTTAACTTG
901 TTTATTGCAG CTTATAATGG TTACAAATAA AGCAATAGCA TCACAAATTT CACAAATAAA
961 GCATTTTTTT CACTGCATTC TAGTTGTGGT TTGTCCAAAC TCATCAATGT ATCTTAAGGC
1021 GTAAATTGTA AGCGTTAATA TTTTGTTAAA ATTCGCGTTA AATTTTTGTT AAATCAGCTC
1081 ATTTTTTAAC CAATAGGCCG AAATCGGCAA AATCCCTTAT AAATCAAAAG AATAGACCGA
1141 GATAGGGTTG AGTGTTGTTC CAGTTTGGAA CAAGAGTCCA CTATTAAAGA ACGTGGACTC
1201 CAACGTCAAA GGGCGAAAAA CCGTCTATCA GGGCGATGGC CCACTACGTG AACCATCACC
1261 CTAATCAAGT TTTTTGGGGT CGAGGTGCCG TAAAGCACTA AATCGGAACC CTAAAGGGAG
1321 CCCCCGATTT AGAGCTTGAC GGGGAAAGCC GGCGAACGTG GCGAGAAAGG AAGGGAAGAA
1381 AGCGAAAGGA GCGGGCGCTA GGGCGCTGGC AAGTGTAGCG GTCACGCTGC GCGTAACCAC
1441 CACACCCGCC GCGCTTAATG CGCCGCTACA GGGCGCGTCA GGTGGCACTT TTCGGGGAAA
1501 TGTGCGCGGA ACCCCTATTT GTTTATTTTT CTAAATACAT TCAAATATGT ATCCGCTCAT
1561 GAGACAATAA CCCTGATAAA TGCTTCAATA ATATTGAAAA AGGAAGAGTC CTGAGGCGGA
1621 AAGAACCAGC TGTGGAATGT GTGTCAGTTA GGGTGTGGAA AGTCCCCAGG CTCCCCAGCA
1681 GGCAGAAGTA TGCAAAGCAT GCATCTCAAT TAGTCAGCAA CCAGGTGTGG AAAGTCCCCA
1741 GGCTCCCCAG CAGGCAGAAG TATGCAAAGC ATGCATCTCA ATTAGTCAGC AACCATAGTC
1801 CCGCCCCTAA CTCCGCCCAT CCCGCCCCTA ACTCCGCCCA GTTCCGCCCA TTCTCCGCCC
1861 CATGGCTGAC TAATTTTTTT TATTTATGCA GAGGCCGAGG CCGCCTCGGC CTCTGAGCTA
1921 TTCCAGAAGT AGTGAGGAGG CTTTTTTGGA GGCCTAGGCT TTTGCAAAGA TCGATCAAGA
1981 GACAGGATGA GGATCGTTTC GCATGATTGA ACAAGATGGA TTGCACGCAG GTTCTCCGGC
2041 CGCTTGGGTG GAGAGGCTAT TCGGCTATGA CTGGGCACAA CAGACAATCG GCTGCTCTGA
2101 TGCCGCCGTG TTCCGGCTGT CAGCGCAGGG GCGCCCGGTT CTTTTTGTCA AGACCGACCT
2161 GTCCGGTGCC CTGAATGAAC TGCAAGACGA GGCAGCGCGG CTATCGTGGC TGGCCACGAC
2221 GGGCGTTCCT TGCGCAGCTG TGCTCGACGT TGTCACTGAA GCGGGAAGGG ACTGGCTGCT
2281 ATTGGGCGAA GTGCCGGGGC AGGATCTCCT GTCATCTCAC CTTGCTCCTG CCGAGAAAGT
2341 ATCCATCATG GCTGATGCAA TGCGGCGGCT GCATACGCTT GATCCGGCTA CCTGCCCATT
2401 CGACCACCAA GCGAAACATC GCATCGAGCG AGCACGTACT CGGATGGAAG CCGGTCTTGT
2461 CGATCAGGAT GATCTGGACG AAGAGCATCA GGGGCTCGCG CCAGCCGAAC TGTTCGCCAG
2521 GCTCAAGGCG AGCATGCCCG ACGGCGAGGA TCTCGTCGTG ACCCATGGCG ATGCCTGCTT
2581 GCCGAATATC ATGGTGGAAA ATGGCCGCTT TTCTGGATTC ATCGACTGTG GCCGGCTGGG
2641 TGTGGCGGAC CGCTATCAGG ACATAGCGTT GGCTACCCGT GATATTGCTG AAGAGCTTGG
2701 CGGCGAATGG GCTGACCGCT TCCTCGTGCT TTACGGTATC GCCGCTCCCG ATTCGCAGCG
2761 CATCGCCTTC TATCGCCTTC TTGACGAGTT CTTCTGAGCG GGACTCTGGG GTTCGAAATG
2821 ACCGACCAAG CGACGCCCAA CCTGCCATCA CGAGATTTCG ATTCCACCGC CGCCTTCTAT
2881 GAAAGGTTGG GCTTCGGAAT CGTTTTCCGG GACGCCGGCT GGATGATCCT CCAGCGCGGG
2941 GATCTCATGC TGGAGTTCTT CGCCCACCCT AGGGGGAGGC TAACTGAAAC ACGGAAGGAG
3001 ACAATACCGG AAGGAACCCG CGCTATGACG GCAATAAAAA GACAGAATAA AACGCACGGT
3061 GTTGGGTCGT TTGTTCATAA ACGCGGGGTT CGGTCCCAGG GCTGGCACTC TGTCGATACC
3121 CCACCGAGAC CCCATTGGGG CCAATACGCC CGCGTTTCTT CCTTTTCCCC ACCCCACCCC
3181 CCAAGTTCGG GTGAAGGCCC AGGGCTCGCA GCCAACGTCG GGGCGGCAGG CCCTGCCATA
3241 GCCTCAGGTT ACTCATATAT ACTTTAGATT GATTTAAAAC TTCATTTTTA ATTTAAAAGG
3301 ATCTAGGTGA AGATCCTTTT TGATAATCTC ATGACCAAAA TCCCTTAACG TGAGTTTTCG
3361 TTCCACTGAG CGTCAGACCC CGTAGAAAAG ATCAAAGGAT CTTCTTGAGA TCCTTTTTTT
3421 CTGCGCGTAA TCTGCTGCTT GCAAACAAAA AAACCACCGC TACCAGCGGT GGTTTGTTTG
3481 CCGGATCAAG AGCTACCAAC TCTTTTTCCG AAGGTAACTG GCTTCAGCAG AGCGCAGATA
3541 CCAAATACTG TCCTTCTAGT GTAGCCGTAG TTAGGCCACC ACTTCAAGAA CTCTGTAGCA
3601 CCGCCTACAT ACCTCGCTCT GCTAATCCTG TTACCAGTGG CTGCTGCCAG TGGCGATAAG
3661 TCGTGTCTTA CCGGGTTGGA CTCAAGACGA TAGTTACCGG ATAAGGCGCA GCGGTCGGGC
3721 TGAACGGGGG GTTCGTGCAC ACAGCCCAGC TTGGAGCGAA CGACCTACAC CGAACTGAGA
3781 TACCTACAGC GTGAGCTATG AGAAAGCGCC ACGCTTCCCG AAGGGAGAAA GGCGGACAGG
3841 TATCCGGTAA GCGGCAGGGT CGGAACAGGA GAGCGCACGA GGGAGCTTCC AGGGGGAAAC
3901 GCCTGGTATC TTTATAGTCC TGTCGGGTTT CGCCACCTCT GACTTGAGCG TCGATTTTTG
3961 TGATGCTCGT CAGGGGGGCG GAGCCTATGG AAAAACGCCA GCAACGCGGC CTTTTTACGG
4021 TTCCTGGCCT TTTGCTGGCC TTTTGCTCAC ATGTTCTTTC CTGCGTTATC CCCTGATTCT
4081 GTGGATAACC GTATTACCGC CATGCAT
//
pDsRed2-1载体其他相关的其他类型载体: